Physical Activity Associated Proteomics of Skeletal Muscle: Being Physically Active in Daily Life May Protect Skeletal Muscle From Aging
- PMID: 30971946
- PMCID: PMC6443906
- DOI: 10.3389/fphys.2019.00312
Physical Activity Associated Proteomics of Skeletal Muscle: Being Physically Active in Daily Life May Protect Skeletal Muscle From Aging
Abstract
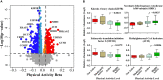
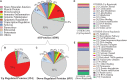
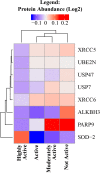
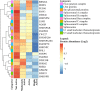
Muscle strength declines with aging and increasing physical activity is the only intervention known to attenuate this decline. In order to adequately investigate both preventive and therapeutic interventions against sarcopenia, a better understanding of the biological changes that are induced by physical activity in skeletal muscle is required. To determine the effect of physical activity on the skeletal muscle proteome, we utilized liquid-chromatography mass spectrometry to obtain quantitative proteomics data on human skeletal muscle biopsies from 60 well-characterized healthy individuals (20-87 years) who reported heterogeneous levels of physical activity (not active, active, moderately active, and highly active). Over 4,000 proteins were quantified, and higher self-reported physical activity was associated with substantial overrepresentation of proteins associated with mitochondria, TCA cycle, structural and contractile muscle, and genome maintenance. Conversely, proteins related to the spliceosome, transcription regulation, immune function, and apoptosis, DNA damage, and senescence were underrepresented with higher self-reported activity. These differences in observed protein expression were related to different levels of physical activity in daily life and not intense competitive exercise. In most instances, differences in protein levels were directly opposite to those reported in the literature observed with aging. These data suggest that being physically active in daily life has strong and biologically detectable beneficial effects on muscle.
Keywords: DNA damage; immunity; mass spectrometry; mitochondria; physical activity; proteomics; skeletal muscle; spliceosome.
Figures

Similar articles
-
Discovery proteomics in aging human skeletal muscle finds change in spliceosome, immunity, proteostasis and mitochondria.Elife. 2019 Oct 23;8:e49874. doi: 10.7554/eLife.49874. Elife. 2019. PMID: 31642809 Free PMC article.
-
Unbiased proteomics, histochemistry, and mitochondrial DNA copy number reveal better mitochondrial health in muscle of high-functioning octogenarians.Elife. 2022 Apr 11;11:e74335. doi: 10.7554/eLife.74335. Elife. 2022. PMID: 35404238 Free PMC article.
-
Combination of exercise training and resveratrol attenuates obese sarcopenia in skeletal muscle atrophy.Chin J Physiol. 2020 May-Jun;63(3):101-112. doi: 10.4103/CJP.CJP_95_19. Chin J Physiol. 2020. PMID: 32594063
-
The pleiotropic effect of physical exercise on mitochondrial dynamics in aging skeletal muscle.Oxid Med Cell Longev. 2015;2015:917085. doi: 10.1155/2015/917085. Epub 2015 Apr 5. Oxid Med Cell Longev. 2015. PMID: 25945152 Free PMC article. Review.
-
Protein and exercise in the prevention of sarcopenia and aging.Nutr Res. 2017 Apr;40:1-20. doi: 10.1016/j.nutres.2017.01.001. Epub 2017 Jan 16. Nutr Res. 2017. PMID: 28473056 Review.
Cited by
-
The Mitochondrial Proteomic Signatures of Human Skeletal Muscle Linked to Insulin Resistance.Int J Mol Sci. 2020 Jul 28;21(15):5374. doi: 10.3390/ijms21155374. Int J Mol Sci. 2020. PMID: 32731645 Free PMC article. Review.
-
Discovery proteomics in aging human skeletal muscle finds change in spliceosome, immunity, proteostasis and mitochondria.Elife. 2019 Oct 23;8:e49874. doi: 10.7554/eLife.49874. Elife. 2019. PMID: 31642809 Free PMC article.
-
The 2022 Report on the Human Proteome from the HUPO Human Proteome Project.J Proteome Res. 2023 Apr 7;22(4):1024-1042. doi: 10.1021/acs.jproteome.2c00498. Epub 2022 Nov 1. J Proteome Res. 2023. PMID: 36318223 Free PMC article. Review.
-
Cross-Sectional Associations between Clinical Biochemistry and Nutritional Biomarkers and Sarcopenic Indices of Skeletal Muscle in the Baltimore Longitudinal Study of Aging.J Nutr. 2025 May;155(5):1535-1548. doi: 10.1016/j.tjnut.2025.03.006. Epub 2025 Mar 8. J Nutr. 2025. PMID: 40064424 Free PMC article.
-
Sca1+ Progenitor Cells (Ex vivo) Exhibits Differential Proteomic Signatures From the Culture Adapted Sca1+ Cells (In vitro), Both Isolated From Murine Skeletal Muscle Tissue.Stem Cell Rev Rep. 2021 Oct;17(5):1754-1767. doi: 10.1007/s12015-021-10134-w. Epub 2021 Mar 19. Stem Cell Rev Rep. 2021. PMID: 33742350
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

