Plant microRNAs: Biogenesis, Homeostasis, and Degradation
- PMID: 30972093
- PMCID: PMC6445950
- DOI: 10.3389/fpls.2019.00360
Plant microRNAs: Biogenesis, Homeostasis, and Degradation
Abstract
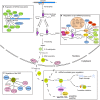
MicroRNAs (miRNAs), a class of endogenous, tiny, non-coding RNAs, are master regulators of gene expression among most eukaryotes. Intracellular miRNA abundance is regulated under multiple levels of control including transcription, processing, RNA modification, RNA-induced silencing complex (RISC) assembly, miRNA-target interaction, and turnover. In this review, we summarize our current understanding of the molecular components and mechanisms that influence miRNA biogenesis, homeostasis, and degradation in plants. We also make comparisons with findings from other organisms where necessary.
Keywords: Argonaute; DCL1; HEN1; miRNA biogenesis; target mimic; uridylation.
Figures

References
-
- Achkar N. P., Cho S. K., Poulsen C., Arce A. L., Re D. A., Giudicatti A. J., et al. (2018). A Quick HYL1-dependent reactivation of MicroRNA production is required for a proper developmental response after extended periods of light deprivation. Dev Cell 46 236–247. 10.1016/j.devcel.2018.06.014 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

