Urolithin A-activated autophagy but not mitophagy protects against ischemic neuronal injury by inhibiting ER stress in vitro and in vivo
- PMID: 30972969
- PMCID: PMC6698978
- DOI: 10.1111/cns.13136
Urolithin A-activated autophagy but not mitophagy protects against ischemic neuronal injury by inhibiting ER stress in vitro and in vivo
Abstract
Aim: Mitochondrial autophagy (mitophagy) clears damaged mitochondria and attenuates ischemic neuronal injury. Urolithin A (Uro-A) activates mitophagy in mammal cells and Caenorhabditis elegans. We explored neuroprotection of Uro-A against ischemic neuronal injury.
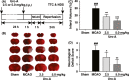
Methods: Mice were subjected to middle cerebral artery occlusion. The brain infarct and neurological deficit scores were measured. The N2a cells and primary cultured mice cortical neurons were subjected to oxygen-glucose deprivation and reperfusion (OGD/R). Uro-A was incubated during OGD/R, and cell injury was determined by MTT and LDH. Autophagosomes were visualized by transfecting mCherry-microtubule-associated protein 1 light chain 3 (LC3). The protein levels of LC3-II, p62, Translocase Of Inner Mitochondrial Membrane 23 (TIMM23), and cytochrome c oxidase subunit 4 isoform 1 (COX4I1) were detected by Western blot. The ER stress markers, activating transcription factor 6 (ATF6) and C/EBP homologous protein (CHOP), were determined by reverse transcription-polymerase chain reaction (RT-PCR).
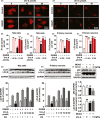
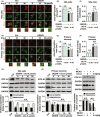
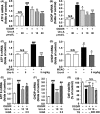
Results: Urolithin A alleviated OGD/R-induced injury in N2a cells and neurons and reduced ischemic brain injury in mice. Uro-A reinforced ischemia-induced autophagy. Furthermore, Uro-A-conferred protection was abolished by 3-methyladenine, suggesting the requirement of autophagy for neuroprotection. However, mitophagy was not further activated by Uro-A. Instead, Uro-A attenuated OGD/R-induced ER stress, which was abolished by 3-methyladenosine. Additionally, neuroprotection was reversed by ER stress inducer.
Conclusion: Urolithin A protected against ischemic neuronal injury by reinforcing autophagy rather than mitophagy. Autophagy activation by Uro-A attenuated ischemic neuronal death by suppressing ER stress.
Keywords: autophagy/mitophagy; cerebral ischemia; endoplasmic reticulum stress; neuroprotection; urolithin A.
© 2019 The Authors. CNS Neuroscience & Therapeutics Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest.
Figures


References
-
- Feigin VL, Norrving B, Mensah GA. Global burden of stroke. Circ Res. 2017;120(3):439‐448. - PubMed
-
- Dirnagl U, Endres M. Found in translation: preclinical stroke research predicts human pathophysiology, clinical phenotypes, and therapeutic outcomes. Stroke. 2014;45(5):1510‐1518. - PubMed
-
- Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19(6):349‐364. - PubMed
-
- Gabryel B, Kost A, Kasprowska D. Neuronal autophagy in cerebral ischemia–a potential target for neuroprotective strategies? Pharmacol Rep. 2012;64(1):1‐15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

