The all-intracellular order Legionellales is unexpectedly diverse, globally distributed and lowly abundant
- PMID: 30973601
- PMCID: PMC6167759
- DOI: 10.1093/femsec/fiy185
The all-intracellular order Legionellales is unexpectedly diverse, globally distributed and lowly abundant
Abstract
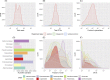
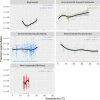
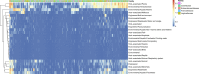
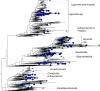
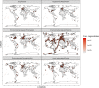
Legionellales is an order of the Gammaproteobacteria, only composed of host-adapted, intracellular bacteria, including the accidental human pathogens Legionella pneumophila and Coxiella burnetii. Although the diversity in terms of lifestyle is large across the order, only a few genera have been sequenced, owing to the difficulty to grow intracellular bacteria in pure culture. In particular, we know little about their global distribution and abundance. Here, we analyze 16/18S rDNA amplicons both from tens of thousands of published studies and from two separate sampling campaigns in and around ponds and in a silver mine. We demonstrate that the diversity of the order is much larger than previously thought, with over 450 uncultured genera. We show that Legionellales are found in about half of the samples from freshwater, soil and marine environments and quasi-ubiquitous in man-made environments. Their abundance is low, typically 0.1%, with few samples up to 1%. Most Legionellales OTUs are globally distributed, while many do not belong to a previously identified species. This study sheds a new light on the ubiquity and diversity of one major group of host-adapted bacteria. It also emphasizes the need to use metagenomics to better understand the role of host-adapted bacteria in all environments.
Keywords: amplicons; geographical distribution; host-adapted bacteria; legionella; legionellales; metagenomics.
Figures

References
-
- Andrews S. FastQC. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ 2010.
-
- Bouchon D, Cordaux R, Grève P. Rickettsiella, intracellular pathogens of Arthropods. Manipulative Tenants: Bacteria associated with Arthropods, In: Zchori-Fein E, Bourtzis K, (eds). pp. 127–45.. CRC Press, Boca Raton, FL; 2011.
-
- Carvalho FRS, Nastasi FR, Gamba RC et al. Occurrence and diversity of legionellaceae in polar lakes of the antarctic peninsula. Curr Microbiol. 2008;57:294–300. - PubMed

