Detection of circulating extracellular mRNAs by modified small-RNA-sequencing analysis
- PMID: 30973829
- PMCID: PMC6538341
- DOI: 10.1172/jci.insight.127317
Detection of circulating extracellular mRNAs by modified small-RNA-sequencing analysis
Abstract
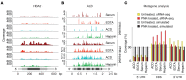
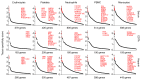
Extracellular mRNAs (ex-mRNAs) potentially supersede extracellular miRNAs (ex-miRNAs) and other RNA classes as biomarkers. We performed conventional small-RNA-sequencing (sRNA-seq) and sRNA-seq with T4 polynucleotide kinase (PNK) end-treatment of total exRNA isolated from serum and platelet-poor EDTA, ACD, and heparin plasma to study the effect on ex-mRNA capture. Compared to conventional sRNA-seq PNK-treatment increased the detection of informative ex-mRNAs reads up to 50-fold. The exRNA pool was dominated by hematopoietic cells and platelets, with additional contribution from the liver. About 60% of the 15- to 42-nt reads originated from the coding sequences, in a pattern reminiscent of ribosome-profiling. Blood sample type had a considerable influence on the exRNA profile. On average approximately 350 to 1,100 distinct ex-mRNA transcripts were detected depending on plasma type. In serum, additional transcripts from neutrophils and hematopoietic cells increased this number to near 2,300. EDTA and ACD plasma showed a destabilizing effect on ex mRNA and non-coding RNA ribonucleoprotein complexes compared to other plasma types. In a proof-of-concept study, we investigated differences between the exRNA profiles of patients with acute coronary syndrome (ACS) and healthy controls. The improved tissue resolution of ex mRNAs after PNK-treatment enabled us to detect a neutrophil-signature in ACS that escaped detection by ex miRNA analysis.
Keywords: Bioinformatics; Cardiology; Molecular diagnosis; RNA processing; Vascular Biology.
Conflict of interest statement
Figures



References
-
- Mandel P, Metais P. [Not Available] C R Seances Soc Biol Fil. 1948;142(3-4):241–243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

