Immunoproteomic Lessons for Human Respiratory Syncytial Virus Vaccine Design
- PMID: 30974886
- PMCID: PMC6518116
- DOI: 10.3390/jcm8040486
Immunoproteomic Lessons for Human Respiratory Syncytial Virus Vaccine Design
Abstract
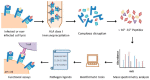
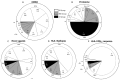
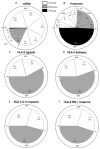
Accurate antiviral humoral and cellular immune responses require prior recognition of antigenic peptides presented by human leukocyte antigen (HLA) class I and II molecules on the surface of antigen-presenting cells. Both the helper and the cytotoxic immune responses are critical for the control and the clearance of human respiratory syncytial virus (HRSV) infection, which is a significant cause of morbidity and mortality in infected pediatric, immunocompromised and elderly populations. In this article we review the immunoproteomics studies which have defined the general antigen processing and presentation rules that determine both the immunoprevalence and the immunodominance of the cellular immune response to HRSV. Mass spectrometry and functional analyses have shown that the HLA class I and II cellular immune responses against HRSV are mainly focused on three viral proteins: fusion, matrix, and nucleoprotein. Thus, these studies have important implications for vaccine development against this virus, since a vaccine construct including these three relevant HRSV proteins could efficiently stimulate the major components of the adaptive immune system: humoral, helper, and cytotoxic effector immune responses.
Keywords: Antigen processing; HLA; T cells; immune response; immunoproteomics; mass spectrometry; respiratory infectious disease; vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Collins P.L., Chanock R.M., Murphy B.R. Respiratory Syncytial Virus. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2013. pp. 1086–1124.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

