Proliferation of hydrocarbon-degrading microbes at the bottom of the Mariana Trench
- PMID: 30975208
- PMCID: PMC6460516
- DOI: 10.1186/s40168-019-0652-3
Proliferation of hydrocarbon-degrading microbes at the bottom of the Mariana Trench
Abstract
Background: The Mariana Trench is the deepest known site in the Earth's oceans, reaching a depth of ~ 11,000 m at the Challenger Deep. Recent studies reveal that hadal waters harbor distinctive microbial planktonic communities. However, the genetic potential of microbial communities within the hadal zone is poorly understood.
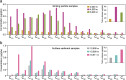
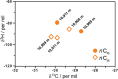
Results: Here, implementing both culture-dependent and culture-independent methods, we perform extensive analysis of microbial populations and their genetic potential at different depths in the Mariana Trench. Unexpectedly, we observed an abrupt increase in the abundance of hydrocarbon-degrading bacteria at depths > 10,400 m in the Challenger Deep. Indeed, the proportion of hydrocarbon-degrading bacteria at > 10,400 m is the highest observed in any natural environment on Earth. These bacteria were mainly Oleibacter, Thalassolituus, and Alcanivorax genera, all of which include species known to consume aliphatic hydrocarbons. This community shift towards hydrocarbon degraders was accompanied by increased abundance and transcription of genes involved in alkane degradation. Correspondingly, three Alcanivorax species that were isolated from 10,400 m water supplemented with hexadecane were able to efficiently degrade n-alkanes under conditions simulating the deep sea, as did a reference Oleibacter strain cultured at atmospheric pressure. Abundant n-alkanes were observed in sinking particles at 2000, 4000, and 6000 m (averaged 23.5 μg/gdw) and hadal surface sediments at depths of 10,908, 10,909, and 10,911 m (averaged 2.3 μg/gdw). The δ2H values of n-C16/18 alkanes that dominated surface sediments at near 11,000-m depths ranged from - 79 to - 93‰, suggesting that these sedimentary alkanes may have been derived from an unknown heterotrophic source.
Conclusions: These results reveal that hydrocarbon-degrading microorganisms are present in great abundance in the deepest seawater on Earth and shed a new light on potential biological processes in this extreme environment.
Keywords: Challenger Deep; Hadal water; Hydrocarbon biosynthesis; Hydrocarbon degradation; Mariana Trench; Metagenomics; Microbial community.
Conflict of interest statement
Ethics and consent for publication
To carry out sampling, the authors received a ‘Research and Training Permit for the Exclusive Economic Zone of the Federated States of Micronesia’ issued by the National Oceanic Resource Management Authority - Federated States of Micronesia.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
-
- Tian J, Fan L, Liu H, Liu J, Li Y, Qin Q, et al. A nearly uniform distributional pattern of heterotrophic bacteria in the Mariana Trench interior. Deep Sea Res Part 1 Oceanogr Res Pap. 2018;142:116–26.
-
- Nunoura T, Hirai M, Yoshida-Takashima Y, Nishizawa M, Kawagucci S, Yokokawa T, et al. Distribution and niche separation of planktonic microbial communities in the water columns from the surface to the hadal waters of the Japan Trench under the Eutrophic Ocean. Front Microbiol. 2016;7:1261. doi: 10.3389/fmicb.2016.01261. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

