Complete Regression of Advanced Pancreatic Ductal Adenocarcinomas upon Combined Inhibition of EGFR and C-RAF
- PMID: 30975481
- PMCID: PMC10132447
- DOI: 10.1016/j.ccell.2019.03.002
Complete Regression of Advanced Pancreatic Ductal Adenocarcinomas upon Combined Inhibition of EGFR and C-RAF
Abstract
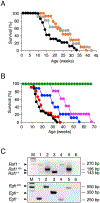
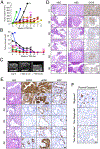
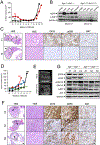
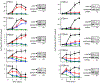
Five-year survival for pancreatic ductal adenocarcinoma (PDAC) patients remains below 7% due to the lack of effective treatments. Here, we report that combined ablation of EGFR and c-RAF expression results in complete regression of a significant percentage of PDAC tumors driven by Kras/Trp53 mutations in genetically engineered mice. Moreover, systemic elimination of these targets induces toxicities that are well tolerated. Response to this targeted therapy correlates with transcriptional profiles that resemble those observed in human PDACs. Finally, inhibition of EGFR and c-RAF expression effectively blocked tumor progression in nine independent patient-derived xenografts carrying KRAS and TP53 mutations. These results open the door to the development of targeted therapies for PDAC patients.
Keywords: Cdk4; EGFR; Erlotinib; PDX tumor models; Pancreatic cancer; c-Raf; therapeutic mouse models; transcriptional profiles; tumor regression.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
M.B. reports a research contract from Pfizer and Eli Lilly and paid consultancy from Amcure.
J.I. reports paid consultancy from Dynasio S.A. and Oncomedics, both in France.
M.H. reports research contracts and/or paid consultancy with Roche and Astra-Zeneca.
None of these relationships are related to the work reported in this manuscript.
We declare a patent application related to this work: EP18382555 Barbacid, M., Guerra C., Blasco, M.T., Navas, C. (2018). COMBINED THERAPY AGAINST CANCER.
Figures




References
-
- Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, Miller DK, Christ AN, Bruxner TJC, Quinn MC, et al. (2016). Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

