Exome array analysis of rare and low frequency variants in amyotrophic lateral sclerosis
- PMID: 30976013
- PMCID: PMC6459905
- DOI: 10.1038/s41598-019-42091-3
Exome array analysis of rare and low frequency variants in amyotrophic lateral sclerosis
Abstract
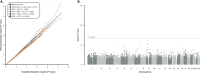
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease that affects 1 in ~350 individuals. Genetic association studies have established ALS as a multifactorial disease with heritability estimated at ~61%, and recent studies show a prominent role for rare variation in its genetic architecture. To identify rare variants associated with disease onset we performed exome array genotyping in 4,244 cases and 3,106 controls from European cohorts. In this largest exome-wide study of rare variants in ALS to date, we performed single-variant association testing, gene-based burden, and exome-wide individual set-unique burden (ISUB) testing to identify single or aggregated rare variation that modifies disease risk. In single-variant testing no variants reached exome-wide significance, likely due to limited statistical power. Gene-based burden testing of rare non-synonymous and loss-of-function variants showed NEK1 as the top associated gene. ISUB analysis did not show an increased exome-wide burden of deleterious variants in patients, possibly suggesting a more region-specific role for rare variation. Complete summary statistics are released publicly. This study did not implicate new risk loci, emphasizing the immediate need for future large-scale collaborations in ALS that will expand available sample sizes, increase genome coverage, and improve our ability to detect rare variants associated to ALS.
Conflict of interest statement
L.H. van den Berg serves on scientific advisory boards for the Prinses Beatrix Spierfonds, Thierry Latran Foundation, Biogen and Cytokinetics; and serves on the editorial board of Amyotrophic Lateral Sclerosis And Frontotemporal Degeneration and The Journal of Neurology, Neurosurgery, and Psychiatry. J.H. Veldink reports that his institute received consultancy fees from Vertex Pharmaceuticals outside the submitted work. O. Hardiman has received speaking honoraria from Novarits, Biogen Idec, Sanofi Aventis and Merck-Serono, has been a member of advisory panels for Biogen Idec, Allergen, Ono Pharmaceuticals, Novartis, Cytokinetics and Sanofi Aventis and serves as Editor-in-Chief of Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Other authors have no reported conflicts of interest.
Figures
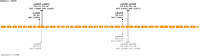
References
-
- Miller RG, M. J. M. D. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neurondisease (MND). The Cochrane Library 1–36 (2012).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

