Nanobody-CD16 Catch Bond Reveals NK Cell Mechanosensitivity
- PMID: 30979550
- PMCID: PMC6486492
- DOI: 10.1016/j.bpj.2019.03.012
Nanobody-CD16 Catch Bond Reveals NK Cell Mechanosensitivity
Abstract
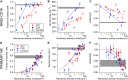
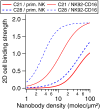
Antibodies are key tools in biomedical research and medicine. Their binding properties are classically measured in solution and characterized by an affinity. However, in physiological conditions, antibodies can bridge an immune effector cell and an antigen-presenting cell, implying that mechanical forces may apply to the bonds. For example, in antibody-dependent cell cytotoxicity-a major mode of action of therapeutic monoclonal antibodies-the Fab domains bind the antigens on the target cell, whereas the Fc domain binds to the activating receptor CD16 (also known as FcgRIII) of an immune effector cell, in a quasi-bidimensional environment (2D). Therefore, there is a strong need to investigate antigen/antibody binding under force (2D) to better understand and predict antibody activity in vivo. We used two anti-CD16 nanobodies targeting two different epitopes and laminar flow chamber assay to measure the association and dissociation of single bonds formed between microsphere-bound CD16 antigens and surface-bound anti-CD16 nanobodies (or single-domain antibodies), simulating 2D encounters. The two nanobodies exhibit similar 2D association kinetics, characterized by a strong dependence on the molecular encounter duration. However, their 2D dissociation kinetics strongly differ as a function of applied force: one exhibits a slip bond behavior in which off rate increases with force, and the other exhibits a catch-bond behavior in which off rate decreases with force. This is the first time, to our knowledge, that catch-bond behavior was reported for antigen-antibody bond. Quantification of natural killer cells spreading on surfaces coated with the nanobodies provides a comparison between 2D and three-dimensional adhesion in a cellular context, supporting the hypothesis of natural killer cell mechanosensitivity. Our results may also have strong implications for the design of efficient bispecific antibodies for therapeutic applications.
Copyright © 2019 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Murphy K., Travers P., Walport M. Seventh Edition. Garland Science; New York, NY: 2008. Janeway’s Immunobiology.
-
- Victora G.D., Nussenzweig M.C. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. - PubMed
-
- Tolar P., Spillane K.M. Force generation in B-cell synapses: mechanisms coupling B-cell receptor binding to antigen internalization and affinity discrimination. Adv. Immunol. 2014;123:69–100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

