Polyploidy spectrum: a new marker in HCC classification
- PMID: 30979717
- PMCID: PMC6984053
- DOI: 10.1136/gutjnl-2018-318021
Polyploidy spectrum: a new marker in HCC classification
Abstract
Objectives: Polyploidy is a fascinating characteristic of liver parenchyma. Hepatocyte polyploidy depends on the DNA content of each nucleus (nuclear ploidy) and the number of nuclei per cell (cellular ploidy). Which role can be assigned to polyploidy during human hepatocellular carcinoma (HCC) development is still an open question. Here, we investigated whether a specific ploidy spectrum is associated with clinical and molecular features of HCC.
Design: Ploidy spectra were determined on surgically resected tissues from patients with HCC as well as healthy control tissues. To define ploidy profiles, a quantitative and qualitative in situ imaging approach was used on paraffin tissue liver sections.
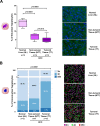
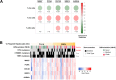
Results: We first demonstrated that polyploid hepatocytes are the major components of human liver parenchyma, polyploidy being mainly cellular (binuclear hepatocytes). Across liver lobules, polyploid hepatocytes do not exhibit a specific zonation pattern. During liver tumorigenesis, cellular ploidy is drastically reduced; binuclear polyploid hepatocytes are barely present in HCC tumours. Remarkably, nuclear ploidy is specifically amplified in HCC tumours. In fact, nuclear ploidy is amplified in HCCs harbouring a low degree of differentiation and TP53 mutations. Finally, our results demonstrated that highly polyploid tumours are associated with a poor prognosis.
Conclusions: Our results underline the importance of quantification of cellular and nuclear ploidy spectra during HCC tumorigenesis.
Keywords: cell cycle; hepatocellular carcinoma; histopathology; liver; molecular pathology.
© Author(s) (or their employer(s)) 2020. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
-
- Ganem NJ, Storchova Z, Tetraploidy PD. aneuploidy and cancer. Curr Opin Genet Dev 2007;17:157–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
