Machine learning-assisted directed protein evolution with combinatorial libraries
- PMID: 30979809
- PMCID: PMC6500146
- DOI: 10.1073/pnas.1901979116
Machine learning-assisted directed protein evolution with combinatorial libraries
Erratum in
-
Correction for Wu et al., Machine learning-assisted directed protein evolution with combinatorial libraries.Proc Natl Acad Sci U S A. 2020 Jan 7;117(1):788-789. doi: 10.1073/pnas.1921770117. Epub 2019 Dec 30. Proc Natl Acad Sci U S A. 2020. PMID: 31888994 Free PMC article. No abstract available.
Abstract
To reduce experimental effort associated with directed protein evolution and to explore the sequence space encoded by mutating multiple positions simultaneously, we incorporate machine learning into the directed evolution workflow. Combinatorial sequence space can be quite expensive to sample experimentally, but machine-learning models trained on tested variants provide a fast method for testing sequence space computationally. We validated this approach on a large published empirical fitness landscape for human GB1 binding protein, demonstrating that machine learning-guided directed evolution finds variants with higher fitness than those found by other directed evolution approaches. We then provide an example application in evolving an enzyme to produce each of the two possible product enantiomers (i.e., stereodivergence) of a new-to-nature carbene Si-H insertion reaction. The approach predicted libraries enriched in functional enzymes and fixed seven mutations in two rounds of evolution to identify variants for selective catalysis with 93% and 79% ee (enantiomeric excess). By greatly increasing throughput with in silico modeling, machine learning enhances the quality and diversity of sequence solutions for a protein engineering problem.
Keywords: catalysis; directed evolution; enzyme; machine learning; protein engineering.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
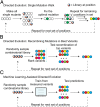


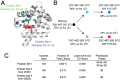

References
-
- Petrović D, Kamerlin SCL. Molecular modeling of conformational dynamics and its role in enzyme evolution. Curr Opin Struct Biol. 2018;52:50–57. - PubMed
-
- Goldsmith M, Tawfik DS. Enzyme engineering: Reaching the maximal catalytic efficiency peak. Curr Opin Struct Biol. 2017;47:140–150. - PubMed
-
- Zeymer C, Hilvert D. Directed evolution of protein catalysts. Annu Rev Biochem. 2018;87:131–157. - PubMed
-
- Garcia-Borrás M, Houk KN, Jiménez-Oses G. Computational design of protein function. In: Martín-Santamaría S, editor. Computational Tools for Chemical Biology. Royal Society of Chemistry; London: 2018. pp. 87–107.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

