Encrustations on ureteral stents from patients without urinary tract infection reveal distinct urotypes and a low bacterial load
- PMID: 30981280
- PMCID: PMC6462311
- DOI: 10.1186/s40168-019-0674-x
Encrustations on ureteral stents from patients without urinary tract infection reveal distinct urotypes and a low bacterial load
Abstract
Background: Current knowledge of the urinary tract microbiome is limited to urine analysis and analysis of biofilms formed on Foley catheters. Bacterial biofilms on ureteral stents have rarely been investigated, and no cultivation-independent data are available on the microbiome of the encrustations on the stents.
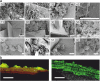
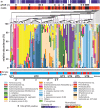
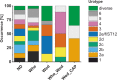
Results: The typical encrustations of organic and inorganic urine-derived material, including microbial biofilms formed during 3-6 weeks on ureteral stents in patients treated for kidney and ureteral stones, and without reported urinary tract infection at the time of stent insertion, were analysed. Next-generation sequencing of the 16S rRNA gene V3-V4 region revealed presence of different urotypes, distinct bacterial communities. Analysis of bacterial load was performed by combining quantification of 16S rRNA gene copy numbers by qPCR with microscopy and cultivation-dependent analysis methods, which revealed that ureteral stent biofilms mostly contain low numbers of bacteria. Fluorescence microscopy indicates the presence of extracellular DNA. Bacteria identified in biofilms by microscopy had mostly morphogenic similarities to gram-positive bacteria, in few cases to Lactobacillus and Corynebacterium, while sequencing showed many additional bacterial genera. Weddellite crystals were absent in biofilms of patients with Enterobacterales and Corynebacterium-dominated microbiomes.
Conclusions: This study provides novel insights into the bacterial burden in ureteral stent encrustations and the urinary tract microbiome. Short-term (3-6 weeks) ureteral stenting is associated with a low load of viable and visible bacteria in ureteral stent encrustations, which may be different from long-term stenting. Patients could be classified according to different urotypes, some of which were dominated by potentially pathogenic species. Facultative pathogens however appear to be a common feature in patients without clinically manifested urinary tract infection.
Trial registration: ClinicalTrials.gov, NCT02845726 . Registered on 30 June 2016-retrospectively registered.
Keywords: Biofilm; Cultivation-independent methods; Encrustation; Microbiome; Next-generation sequencing; Ureteral stent; Urinary tract microbiota; qPCR.
Conflict of interest statement
Ethics approval and consent to participate
Approval of the study was obtained from local ethics committee (EKSG 15/084). Procedures performed in the study were all in accordance with the ethical standards of the institutional and national research committee, with the 1964 Helsinki Declaration, and its later amendments. All individual participants included in the study had given written informed consent.
Consent for publication
Not applicable
Competing interests
Sebastian Strempel is employee of Microsynth AG.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh M-J, Huang S-T, Ljungberg I, Sprague BM, Lucas SK, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med. 2012;10:174. doi: 10.1186/1479-5876-10-174. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

