Promoter-Intrinsic and Local Chromatin Features Determine Gene Repression in LADs
- PMID: 30982597
- PMCID: PMC6506275
- DOI: 10.1016/j.cell.2019.03.009
Promoter-Intrinsic and Local Chromatin Features Determine Gene Repression in LADs
Abstract
It is largely unclear whether genes that are naturally embedded in lamina-associated domains (LADs) are inactive due to their chromatin environment or whether LADs are merely secondary to the lack of transcription. We show that hundreds of human promoters become active when moved from their native LAD position to a neutral context in the same cells, indicating that LADs form a repressive environment. Another set of promoters inside LADs is able to "escape" repression, although their transcription elongation is attenuated. By inserting reporters into thousands of genomic locations, we demonstrate that escaper promoters are intrinsically less sensitive to LAD repression. This is not simply explained by promoter strength but by the interplay between promoter sequence and local chromatin features that vary strongly across LADs. Enhancers also differ in their sensitivity to LAD chromatin. This work provides a general framework for the systematic understanding of gene regulation by repressive chromatin.
Keywords: GRO-cap; SuRE; chromatin; lamina-associated domains; massive parallel reporter assay; promoters; repression; thousands of reporters integrated in parallel.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




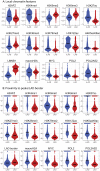
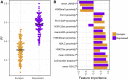
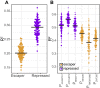


References
-
- Akhtar W., de Jong J., Pindyurin A.V., Pagie L., Meuleman W., de Ridder J., Berns A., Wessels L.F.A., van Lohuizen M., van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. - PubMed
-
- Akhtar W., Pindyurin A.V., de Jong J., Pagie L., Ten Hoeve J., Berns A., Wessels L.F.A., van Steensel B., van Lohuizen M. Using TRIP for genome-wide position effect analysis in cultured cells. Nat. Protoc. 2014;9:1255–1281. - PubMed
-
- Amendola M., Venneri M.A., Biffi A., Vigna E., Naldini L. Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nat. Biotechnol. 2005;23:108–116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

