A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer
- PMID: 30982598
- PMCID: PMC6526772
- DOI: 10.1016/j.cell.2019.03.005
A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer
Abstract
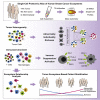
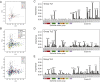
Breast cancer is a heterogeneous disease. Tumor cells and associated healthy cells form ecosystems that determine disease progression and response to therapy. To characterize features of breast cancer ecosystems and their associations with clinical data, we analyzed 144 human breast tumor and 50 non-tumor tissue samples using mass cytometry. The expression of 73 proteins in 26 million cells was evaluated using tumor and immune cell-centric antibody panels. Tumors displayed individuality in tumor cell composition, including phenotypic abnormalities and phenotype dominance. Relationship analyses between tumor and immune cells revealed characteristics of ecosystems related to immunosuppression and poor prognosis. High frequencies of PD-L1+ tumor-associated macrophages and exhausted T cells were found in high-grade ER+ and ER- tumors. This large-scale, single-cell atlas deepens our understanding of breast tumor ecosystems and suggests that ecosystem-based patient classification will facilitate identification of individuals for precision medicine approaches targeting the tumor and its immunoenvironment.
Keywords: T cell; breast cancer; immunosuppression; macrophage; mass cytometry; single-cell analysis; tumor ecosystem; tumor heterogeneity.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures











References
-
- American Joint Committee on Cancer . In: AJCC Cancer Staging Manual. Eighth Edition. Amin M.B., Edge S., Greene F., Byrd D.R., Brookland R.K., Washington M.K., Gershenwald J.E., Compton C.C., Hess K.R., Sullivan D.C., Jessup J.M., Brierley J.D., Gaspar L.E., Schilsky R.L., Balch C.M., Winchester D.P., Asare E.A., Madera M., Gress D.M., Meyer L.R., editors. Springer International Publishing; 2017.
-
- Bandura D.R., Baranov V.I., Ornatsky O.I., Antonov A., Kinach R., Lou X., Pavlov S., Vorobiev S., Dick J.E., Tanner S.D. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

