Targeting adenylate-forming enzymes with designed sulfonyladenosine inhibitors
- PMID: 30982830
- PMCID: PMC6594144
- DOI: 10.1038/s41429-019-0171-2
Targeting adenylate-forming enzymes with designed sulfonyladenosine inhibitors
Abstract
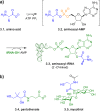
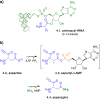
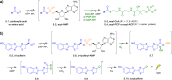
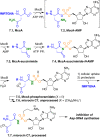
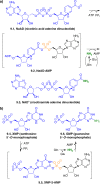
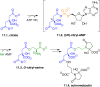
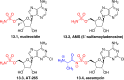
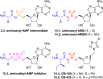
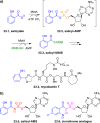
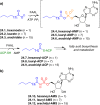
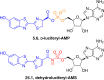
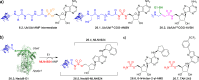
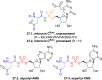
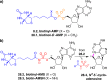
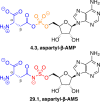
Adenylate-forming enzymes are a mechanistic superfamily that are involved in diverse biochemical pathways. They catalyze ATP-dependent activation of carboxylic acid substrates as reactive acyl adenylate (acyl-AMP) intermediates and subsequent coupling to various nucleophiles to generate ester, thioester, and amide products. Inspired by natural products, acyl sulfonyladenosines (acyl-AMS) that mimic the tightly bound acyl-AMP reaction intermediates have been developed as potent inhibitors of adenylate-forming enzymes. This simple yet powerful inhibitor design platform has provided a wide range of biological probes as well as several therapeutic lead compounds. Herein, we provide an overview of the nine structural classes of adenylate-forming enzymes and examples of acyl-AMS inhibitors that have been developed for each.
Conflict of interest statement
D.S.T. is a coinventor of U.S. Patents 8,461,128 and 8,946,188; International Patent Applications PCT/US2016/055136 and PCT/US2016/055200; and U.S. Provisional Patent Applications 62/527,925, 62/527,932, 62/527,936, 62/527,943, 62/784,323, and 62/802,650 concerning sulfonyladenosine analogues. L.C.S. is a coinventor of U.S. Provisional Patent Application 62/784,323. M.C.L. declares no conflicts of interest.
Figures








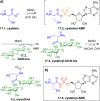

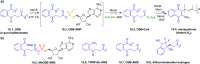
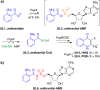

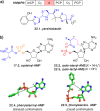
References
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM073546/GM/NIGMS NIH HHS/United States
- R21 AI098802/AI/NIAID NIH HHS/United States
- T32 GM115327/GM/NIGMS NIH HHS/United States
- R33 AI098802/AI/NIAID NIH HHS/United States
- R21 AI063384/AI/NIAID NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 AI118224/AI/NIAID NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
- R01 AI136795/AI/NIAID NIH HHS/United States
- R01 AI075092/AI/NIAID NIH HHS/United States
- F31 AI129244/AI/NIAID NIH HHS/United States
- R01 AI068038/AI/NIAID NIH HHS/United States
- R01 GM100477/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources

