Quantitative Polymerase Chain Reaction Coupled With Sodium Dodecyl Sulfate and Propidium Monoazide for Detection of Viable Streptococcus agalactiae in Milk
- PMID: 30984156
- PMCID: PMC6450196
- DOI: 10.3389/fmicb.2019.00661
Quantitative Polymerase Chain Reaction Coupled With Sodium Dodecyl Sulfate and Propidium Monoazide for Detection of Viable Streptococcus agalactiae in Milk
Abstract
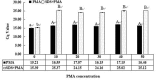
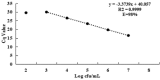
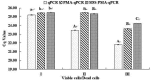
Streptococcus agalactiae is an important pathogen causing bovine mastitis. The aim of this study was to develop a simple and specific method for direct detection of S. agalactiae from milk products. Propidium monoazide (PMA) and sodium dodecyl sulfate (SDS) were utilized to eliminate the interference of dead and injured cells in qPCR. Lysozyme (LYZ) was adopted to increase the extraction efficiency of target bacteria DNA in milk matrix. The specific primers were designed based on cfb gene of S. agalactiae for qPCR. The inclusivity and exclusivity of the assay were evaluated using 30 strains. The method was further determined by the detection of S. agalactiae in spiked milk. Results showed significant differences between the SDS-PMA-qPCR, PMA-qPCR and qPCR when a final concentration of 10 mg/ml (R 2 = 0.9996, E = 95%) of LYZ was added in DNA extraction. Viable S. agalactiae was effectively detected when SDS and PMA concentrations were 20 μg/ml and 10 μM, respectively, and it was specific and more sensitive than qPCR and PMA-qPCR. Moreover, the SDS-PMA-qPCR assay coupled with LYZ was used to detect viable S. agalactiae in spiked milk, with a limit of detection of 3 × 103 cfu/ml. Therefore, the SDS-PMA-qPCR assay had excellent sensitivity and specificity for detection of viable S. agalactiae in milk.
Keywords: Streptococcus agalactiae; milk; propidium monoazide; qPCR; sodium dodecyl sulfate.
Figures


References
-
- Ayman E., Mohamed E., Eman M., Yaser B. (2015). Detection of virulence genes in Staphylococcus aureus and Streptococcus agalactiae isolated from mastitis in the middle east. Br. Microbiol. Res. J. 10 1–9. 10.13140/RG.2.1.4624.8166 - DOI
-
- Boström K. H., Simu K., Hagström K., Riemann L. (2004). Optimization of DNA extraction for quantitative marine bacterioplankton community analysis. Limnol. Oceanogr. Methods 2 365–373. 10.4319/lom.2004.2.365 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

