The Genetic Landscape of Human Glioblastoma and Matched Primary Cancer Stem Cells Reveals Intratumour Similarity and Intertumour Heterogeneity
- PMID: 30984267
- PMCID: PMC6431486
- DOI: 10.1155/2019/2617030
The Genetic Landscape of Human Glioblastoma and Matched Primary Cancer Stem Cells Reveals Intratumour Similarity and Intertumour Heterogeneity
Abstract
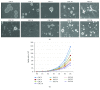
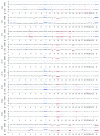
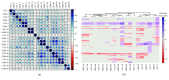
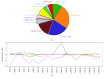
Glioblastoma (GBM) is the most malignant human brain tumour, characterized by rapid progression, invasion, intense angiogenesis, high genomic instability, and resistance to therapies. Despite countless experimental researches for new therapeutic strategies and promising clinical trials, the prognosis remains extremely poor, with a mean survival of less than 14 months. GBM aggressive behaviour is due to a subpopulation of tumourigenic stem-like cells, GBM stem cells (GSCs), which hierarchically drive onset, proliferation, and tumour recurrence. The morbidity and mortality of this disease strongly encourage exploring genetic characteristics of GSCs. Here, using array-CGH platform, we investigated genetic and genomic aberration profiles of GBM parent tumour (n = 10) and their primarily derived GSCs. Statistical analysis was performed by using R software and complex heatmap and corrplot packages. Pearson correlation and K-means algorithm were exploited to compare genetic alterations and to group similar genetic profiles in matched pairs of GBM and derived GSCs. We identified, in both GBM and matched GSCs, recurrent copy number alterations, as chromosome 7 polysomy, chromosome 10 monosomy, and chromosome 9p21deletions, which are typical features of primary GBM, essential for gliomagenesis. These observations suggest a condition of strong genomic instability both in GBM as GSCs. Our findings showed the robust similarity between GBM mass and GSCs (Pearson corr.≥0.65) but also highlighted a marked variability among different patients. Indeed, the heatmap reporting Gain/Loss State for 21022 coding/noncoding genes demonstrated high interpatient divergence. Furthermore, K-means algorithm identified an impairment of pathways related to the development and progression of cancer, such as angiogenesis, as well as pathways related to the immune system regulation, such as T cell activation. Our data confirmed the preservation of the genomic landscape from tumour tissue to GSCs, supporting the relevance of this cellular model to test in vitro new target therapies for GBM.
Figures