An optimized isolation protocol yields high-quality RNA from cassava tissues (Manihot esculenta Crantz)
- PMID: 30984554
- PMCID: PMC6443859
- DOI: 10.1002/2211-5463.12561
An optimized isolation protocol yields high-quality RNA from cassava tissues (Manihot esculenta Crantz)
Abstract
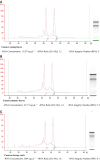
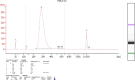
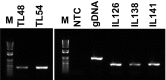
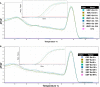
We developed and modified a precise, rapid, and reproducible protocol isolating high-quality RNA from tissues of multiple varieties of cassava plants (Manihot esculenta Crantz). The resulting method is suitable for use in mini, midi, and maxi preparations and rapidly achieves high total RNA yields (170-600 μg·g-1) using low-cost chemicals and consumables and with minimal contamination from polysaccharides, polyphenols, proteins, and other secondary metabolites. In particular, A260 : A280 ratios were > 2.0 for RNA from various tissues, and all of the present RNA samples yielded ribosomal integrity number values of greater than six. The resulting high purity and quality of isolated RNA will facilitate downstream applications (quantitative reverse transcriptase-polymerase chain reaction or RNA sequencing) in cassava molecular breeding.
Keywords: RNA isolation; RNA sequencing; cassava; qRT‐PCR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Highly efficient mesophyll protoplast isolation and PEG-mediated transient gene expression for rapid and large-scale gene characterization in cassava (Manihot esculenta Crantz).BMC Biotechnol. 2017 Mar 14;17(1):29. doi: 10.1186/s12896-017-0349-2. BMC Biotechnol. 2017. PMID: 28292294 Free PMC article.
-
Comparative transcriptomics analysis reveals defense mechanisms of Manihot esculenta Crantz against Sri Lanka Cassava MosaicVirus.BMC Genomics. 2024 May 2;25(1):436. doi: 10.1186/s12864-024-10315-0. BMC Genomics. 2024. PMID: 38698332 Free PMC article.
-
Transfer and expression of an artificial storage protein (ASP1) gene in cassava (Manihot esculenta Crantz).Transgenic Res. 2003 Apr;12(2):243-50. doi: 10.1023/a:1022918925882. Transgenic Res. 2003. PMID: 12739891
-
Cassava genetic transformation and its application in breeding.J Integr Plant Biol. 2011 Jul;53(7):552-69. doi: 10.1111/j.1744-7909.2011.01048.x. Epub 2011 Jun 22. J Integr Plant Biol. 2011. PMID: 21564542 Review.
-
Gene flow between cassava, Manihot esculenta Crantz, and wild relatives.Genet Mol Res. 2003 Dec 30;2(4):334-47. Genet Mol Res. 2003. PMID: 15011137 Review.
Cited by
-
Quality control on oil palm RNA samples for efficient genomic downstream applications.J Genomics. 2024 Feb 17;12:35-43. doi: 10.7150/jgen.92209. eCollection 2024. J Genomics. 2024. PMID: 38434105 Free PMC article.
-
Optimized SDS-Based Protocol for High-Quality RNA Extraction from Musa spp.Methods Protoc. 2025 Feb 19;8(1):21. doi: 10.3390/mps8010021. Methods Protoc. 2025. PMID: 39997645 Free PMC article.
-
Genome-wide analyses of cassava Pathogenesis-related (PR) gene families reveal core transcriptome responses to whitefly infestation, salicylic acid and jasmonic acid.BMC Genomics. 2020 Jan 29;21(1):93. doi: 10.1186/s12864-019-6443-1. BMC Genomics. 2020. PMID: 31996126 Free PMC article.
-
Characterization of cassava ORANGE proteins and their capability to increase provitamin A carotenoids accumulation.PLoS One. 2022 Jan 7;17(1):e0262412. doi: 10.1371/journal.pone.0262412. eCollection 2022. PLoS One. 2022. PMID: 34995328 Free PMC article.
-
An optimized nucleic acid isolation protocol for virus diagnostics in cassava (Manihot esculenta Crantz.).MethodsX. 2021 Aug 21;8:101496. doi: 10.1016/j.mex.2021.101496. eCollection 2021. MethodsX. 2021. PMID: 34754767 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

