Understanding Human-Virus Protein-Protein Interactions Using a Human Protein Complex-Based Analysis Framework
- PMID: 30984872
- PMCID: PMC6456672
- DOI: 10.1128/mSystems.00303-18
Understanding Human-Virus Protein-Protein Interactions Using a Human Protein Complex-Based Analysis Framework
Abstract
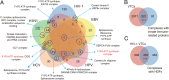
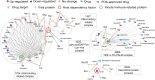
Computational analysis of human-virus protein-protein interaction (PPI) data is an effective way toward systems understanding the molecular mechanism of viral infection. Previous work has mainly focused on characterizing the global properties of viral targets within the entire human PPI network. In comparison, how viruses manipulate host local networks (e.g., human protein complexes) has been rarely addressed from a computational perspective. By mainly integrating information about human-virus PPIs, human protein complexes, and gene expression profiles, we performed a large-scale analysis of virally targeted complexes (VTCs) related to five common human-pathogenic viruses, including influenza A virus subtype H1N1, human immunodeficiency virus type 1, Epstein-Barr virus, human papillomavirus, and hepatitis C virus. We found that viral targets are enriched within human protein complexes. We observed in the context of VTCs that viral targets tended to have a high within-complex degree and to be scaffold and housekeeping proteins. Complexes that are essential for viral propagation were simultaneously targeted by multiple viruses. We characterized the periodic expression patterns of VTCs and provided the corresponding candidates that may be involved in the manipulation of the host cell cycle. As a potential application of the current analysis, we proposed a VTC-based antiviral drug target discovery strategy. Finally, we developed an online VTC-related platform known as VTcomplex (http://zzdlab.com/vtcomplex/index.php or http://systbio.cau.edu.cn/vtcomplex/index.php). We hope that the current analysis can provide new insights into the global landscape of human-virus PPIs at the VTC level and that the developed VTcomplex will become a vital resource for the community. IMPORTANCE Although human protein complexes have been reported to be directly related to viral infection, previous studies have not systematically investigated human-virus PPIs from the perspective of human protein complexes. To the best of our knowledge, we have presented here the most comprehensive and in-depth analysis of human-virus PPIs in the context of VTCs. Our findings confirm that human protein complexes are heavily involved in viral infection. The observed preferences of virally targeted subunits within complexes reflect the mechanisms used by viruses to manipulate host protein complexes. The identified periodic expression patterns of the VTCs and the corresponding candidates could increase our understanding of how viruses manipulate the host cell cycle. Finally, our proposed conceptual application framework of VTCs and the developed VTcomplex could provide new hints to develop antiviral drugs for the clinical treatment of viral infections.
Keywords: antiviral drug discovery; human-virus interaction; network; protein complex; protein-protein interaction.
Figures




References
-
- Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, Hill DE, Vidal M, Kieff E, Johannsen E. 2007. Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci U S A 104:7606–7611. doi: 10.1073/pnas.0702332104. - DOI - PMC - PubMed
-
- de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugue S, Meiffren G, Pradezynski F, Faria BF, Chantier T, Le Breton M, Pellet J, Davoust N, Mangeot PE, Chaboud A, Penin F, Jacob Y, Vidalain PO, Vidal M, Andre P, Rabourdin-Combe C, Lotteau V. 2008. Hepatitis C virus infection protein network. Mol Syst Biol 4:230. doi: 10.1038/msb.2008.66. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Medical
Research Materials

