Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13
- PMID: 30985027
- PMCID: PMC6800999
- DOI: 10.1002/prot.25697
Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13
Abstract
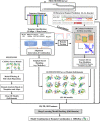
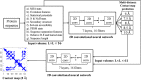
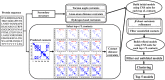
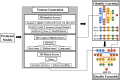
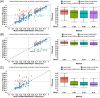
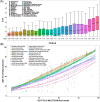
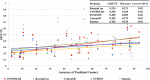
Predicting residue-residue distance relationships (eg, contacts) has become the key direction to advance protein structure prediction since 2014 CASP11 experiment, while deep learning has revolutionized the technology for contact and distance distribution prediction since its debut in 2012 CASP10 experiment. During 2018 CASP13 experiment, we enhanced our MULTICOM protein structure prediction system with three major components: contact distance prediction based on deep convolutional neural networks, distance-driven template-free (ab initio) modeling, and protein model ranking empowered by deep learning and contact prediction. Our experiment demonstrates that contact distance prediction and deep learning methods are the key reasons that MULTICOM was ranked 3rd out of all 98 predictors in both template-free and template-based structure modeling in CASP13. Deep convolutional neural network can utilize global information in pairwise residue-residue features such as coevolution scores to substantially improve contact distance prediction, which played a decisive role in correctly folding some free modeling and hard template-based modeling targets. Deep learning also successfully integrated one-dimensional structural features, two-dimensional contact information, and three-dimensional structural quality scores to improve protein model quality assessment, where the contact prediction was demonstrated to consistently enhance ranking of protein models for the first time. The success of MULTICOM system clearly shows that protein contact distance prediction and model selection driven by deep learning holds the key of solving protein structure prediction problem. However, there are still challenges in accurately predicting protein contact distance when there are few homologous sequences, folding proteins from noisy contact distances, and ranking models of hard targets.
Keywords: contact prediction; deep learning; distance prediction; protein model quality assessment; protein structure prediction; template-based modeling; template-free modeling.
© 2019 The Authors. Proteins: Structure, Function, and Bioinformatics published by Wiley Periodicals, Inc.
Figures



References
-
- Abriata LA, Tamò GE, Monastyrskyy B, Kryshtafovych A, Dal Peraro M. Assessment of hard target modeling in CASP12 reveals an emerging role of alignment‐based contact prediction methods. Proteins. 2018;86:97‐112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

