ChlamBase: a curated model organism database for the Chlamydia research community
- PMID: 30985891
- PMCID: PMC6463448
- DOI: 10.1093/database/baz041
ChlamBase: a curated model organism database for the Chlamydia research community
Erratum in
-
ChlamBase: a curated model organism database for the Chlamydia research community.Database (Oxford). 2019 Jan 1;2019:baz091. doi: 10.1093/database/baz091. Database (Oxford). 2019. PMID: 31211397 Free PMC article. No abstract available.
Abstract
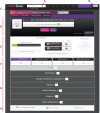
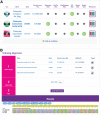
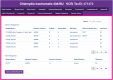
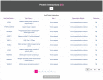
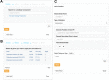
The accelerating growth of genomic and proteomic information for Chlamydia species, coupled with unique biological aspects of these pathogens, necessitates bioinformatic tools and features that are not provided by major public databases. To meet these growing needs, we developed ChlamBase, a model organism database for Chlamydia that is built upon the WikiGenomes application framework, and Wikidata, a community-curated database. ChlamBase was designed to serve as a central access point for genomic and proteomic information for the Chlamydia research community. ChlamBase integrates information from numerous external databases, as well as important data extracted from the literature that are otherwise not available in structured formats that are easy to use. In addition, a key feature of ChlamBase is that it empowers users in the field to contribute new annotations and data as the field advances with continued discoveries. ChlamBase is freely and publicly available at chlambase.org.
© The Author(s) 2019. Published by Oxford University Press.
Figures
References
-
- Miller W.C., Ford C.A., Morris M. et al . (2004) Prevalence of chlamydial and gonococcal infections among young adults in the United States. JAMA, 291, 2229–2236. - PubMed
-
- Stephens R.S., Kalman S., Lammel C. et al . (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science, 282, 754–759. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

