The protective variant rs7173049 at LOXL1 locus impacts on retinoic acid signaling pathway in pseudoexfoliation syndrome
- PMID: 30986821
- PMCID: PMC6644155
- DOI: 10.1093/hmg/ddz075
The protective variant rs7173049 at LOXL1 locus impacts on retinoic acid signaling pathway in pseudoexfoliation syndrome
Abstract
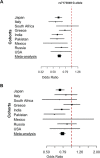
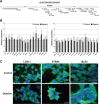
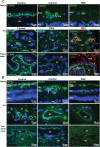
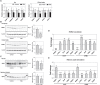
LOXL1 (lysyl oxidase-like 1) has been identified as the major effect locus in pseudoexfoliation (PEX) syndrome, a fibrotic disorder of the extracellular matrix and frequent cause of chronic open-angle glaucoma. However, all known PEX-associated common variants show allele effect reversal in populations of different ancestry, casting doubt on their biological significance. Based on extensive LOXL1 deep sequencing, we report here the identification of a common non-coding sequence variant, rs7173049A>G, located downstream of LOXL1, consistently associated with a decrease in PEX risk (odds ratio, OR = 0.63; P = 6.33 × 10-31) in nine different ethnic populations. We provide experimental evidence for a functional enhancer-like regulatory activity of the genomic region surrounding rs7173049 influencing expression levels of ISLR2 (immunoglobulin superfamily containing leucine-rich repeat protein 2) and STRA6 [stimulated by retinoic acid (RA) receptor 6], apparently mediated by allele-specific binding of the transcription factor thyroid hormone receptor beta. We further show that the protective rs7173049-G allele correlates with increased tissue expression levels of ISLR2 and STRA6 and that both genes are significantly downregulated in tissues of PEX patients together with other key components of the STRA6 receptor-driven RA signaling pathway. siRNA-mediated downregulation of RA signaling induces upregulation of LOXL1 and PEX-associated matrix genes in PEX-relevant cell types. These data indicate that dysregulation of STRA6 and impaired retinoid metabolism are involved in the pathophysiology of PEX syndrome and that the variant rs7173049-G, which represents the first common variant at the broad LOXL1 locus without allele effect reversal, mediates a protective effect through upregulation of STRA6 in ocular tissues.
© The Author(s) 2019. Published by Oxford University Press.
Figures


Similar articles
-
Genotype-correlated expression of lysyl oxidase-like 1 in ocular tissues of patients with pseudoexfoliation syndrome/glaucoma and normal patients.Am J Pathol. 2008 Dec;173(6):1724-35. doi: 10.2353/ajpath.2008.080535. Epub 2008 Oct 30. Am J Pathol. 2008. PMID: 18974306 Free PMC article.
-
Lysyl oxidase-like 1 gene in the reversal of promoter risk allele in pseudoexfoliation syndrome.JAMA Ophthalmol. 2014 Aug;132(8):949-55. doi: 10.1001/jamaophthalmol.2014.845. JAMA Ophthalmol. 2014. PMID: 24809751 Free PMC article.
-
Molecular pathology of pseudoexfoliation syndrome/glaucoma--new insights from LOXL1 gene associations.Exp Eye Res. 2009 Apr;88(4):776-85. doi: 10.1016/j.exer.2008.08.012. Epub 2008 Sep 6. Exp Eye Res. 2009. PMID: 18809397 Review.
-
Association of LOXL1 polymorphisms with pseudoexfoliation, glaucoma, intraocular pressure, and systemic diseases in a Greek population. The Thessaloniki eye study.Invest Ophthalmol Vis Sci. 2014 Jun 10;55(7):4238-43. doi: 10.1167/iovs.14-13991. Invest Ophthalmol Vis Sci. 2014. PMID: 24917141 Free PMC article.
-
Pseudoexfoliation syndrome and glaucoma: from genes to disease mechanisms.Curr Opin Ophthalmol. 2021 Mar 1;32(2):118-128. doi: 10.1097/ICU.0000000000000736. Curr Opin Ophthalmol. 2021. PMID: 33332884 Review.
Cited by
-
Pseudoexfoliative Syndrome in Cataract Surgery-A Quality Register Study and Health Economic Analysis in the Split-Dalmatia County, Croatia.J Clin Med. 2023 Dec 20;13(1):38. doi: 10.3390/jcm13010038. J Clin Med. 2023. PMID: 38202045 Free PMC article.
-
Prevalence of risk alleles in the lysyl oxidase-like 1 gene in pseudoexfoliation glaucoma patients in India.Indian J Ophthalmol. 2022 Jun;70(6):2024-2028. doi: 10.4103/ijo.IJO_2664_21. Indian J Ophthalmol. 2022. PMID: 35647973 Free PMC article.
-
Elevated Intraocular Pressure and Glaucomatous Optic Neuropathy: Genes to Disease Mechanisms, Therapeutic Drugs, and Gene Therapies.Pharmaceuticals (Basel). 2023 Jun 12;16(6):870. doi: 10.3390/ph16060870. Pharmaceuticals (Basel). 2023. PMID: 37375817 Free PMC article. Review.
-
Commentary: The genetics of pseudoexfoliation syndrome/glaucoma.Indian J Ophthalmol. 2022 Jun;70(6):2028-2029. doi: 10.4103/ijo.IJO_30_22. Indian J Ophthalmol. 2022. PMID: 35647974 Free PMC article. No abstract available.
-
Antioxidant Defense and Pseudoexfoliation Syndrome: An Updated Review.Med Sci (Basel). 2022 Dec 14;10(4):68. doi: 10.3390/medsci10040068. Med Sci (Basel). 2022. PMID: 36548003 Free PMC article. Review.
References
-
- Konstas A.G.P. and Ringvold A. (2018) Epidemiology of exfoliation syndrome. J. Glaucoma, 27, S4–S11. - PubMed
-
- Ritch R. and Schlötzer-Schrehardt U. (2001) Exfoliation syndrome. Surv. Ophthalmol., 45, 265–315. - PubMed
-
- Schlötzer-Schrehardt U. and Naumann G.O. (2006) Ocular and systemic pseudoexfoliation syndrome. Am. J. Ophthalmol., 141, 921–937. - PubMed
-
- Ozaki M. (2018) Mechanisms of glaucoma in exfoliation syndrome. J. Glaucoma, 27, S83–S86. - PubMed
-
- Ritch R. (1994) Exfoliation syndrome—the most common identifiable cause of open-angle glaucoma. J. Glaucoma, 3, 176–177. - PubMed

