Characterization of Microbiota Associated with Digesta and Mucosa in Different Regions of Gastrointestinal Tract of Nursery Pigs
- PMID: 30986907
- PMCID: PMC6480377
- DOI: 10.3390/ijms20071630
Characterization of Microbiota Associated with Digesta and Mucosa in Different Regions of Gastrointestinal Tract of Nursery Pigs
Abstract
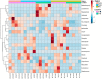
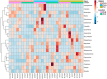
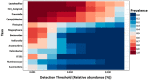
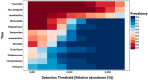
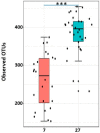
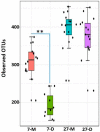
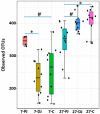
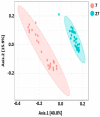
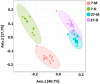
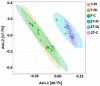
Weaning is a crucial period when piglets have to cope with sudden dietary, social, and environmental stressors that often lead to serious intestinal dysbiosis and mortality. In this study, five mucosal and five digesta samples from each proximate jejunum, distal jejunum, and mid-colon were collected from 7- and 27-day post-weaned pigs and subjected to microbiota analysis using 16S rRNA gene profiling. Taxonomic analysis at phylum level revealed that Proteobacteria was significantly higher at 7 days (13.54%), while Bacteriodetes was higher at 27 days (30.72%) post weaning. Genera such as Campylobacter, Veillonella, Helicobacter, and Blautia that were previously reported in intestinal dysbiosis were significantly enriched in seven-day post-weaned pigs. However, microbial communities shifted as post weaning age increased with a significant increase in alpha diversity, and genera such as Moryella, Dialister, Clostridium, Streptococcus, Prevotella, and Bacteroides become significantly abundant in 27-day post-weaned pigs. Interestingly, the genus Campylobacter was significantly abundant on seven-day post-weaning in two piglets with diarrhea, implicating its role in post-weaning diarrhea. The results of this study suggest that gut microbiota in pigs with dysbiosis on 7-day post weaning undergoes significant changes toward a more normal state as the post-weaning age reaches 27 days.
Keywords: diarrhea; dysbiosis; gut microbiota; pig; post-weaning stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

