Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir
- PMID: 30987343
- PMCID: PMC6520719
- DOI: 10.3390/v11040326
Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir
Abstract
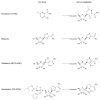
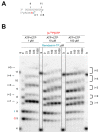
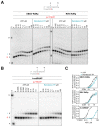
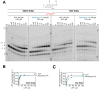
Remdesivir (GS-5734) is a 1'-cyano-substituted adenosine nucleotide analogue prodrug that shows broad-spectrum antiviral activity against several RNA viruses. This compound is currently under clinical development for the treatment of Ebola virus disease (EVD). While antiviral effects have been demonstrated in cell culture and in non-human primates, the mechanism of action of Ebola virus (EBOV) inhibition for remdesivir remains to be fully elucidated. The EBOV RNA-dependent RNA polymerase (RdRp) complex was recently expressed and purified, enabling biochemical studies with the relevant triphosphate (TP) form of remdesivir and its presumptive target. In this study, we confirmed that remdesivir-TP is able to compete for incorporation with adenosine triphosphate (ATP). Enzyme kinetics revealed that EBOV RdRp and respiratory syncytial virus (RSV) RdRp incorporate ATP and remdesivir-TP with similar efficiencies. The selectivity of ATP against remdesivir-TP is ~4 for EBOV RdRp and ~3 for RSV RdRp. In contrast, purified human mitochondrial RNA polymerase (h-mtRNAP) effectively discriminates against remdesivir-TP with a selectivity value of ~500-fold. For EBOV RdRp, the incorporated inhibitor at position i does not affect the ensuing nucleotide incorporation event at position i+1. For RSV RdRp, we measured a ~6-fold inhibition at position i+1 although RNA synthesis was not terminated. Chain termination was in both cases delayed and was seen predominantly at position i+5. This pattern is specific to remdesivir-TP and its 1'-cyano modification. Compounds with modifications at the 2'-position show different patterns of inhibition. While 2'-C-methyl-ATP is not incorporated, ara-ATP acts as a non-obligate chain terminator and prevents nucleotide incorporation at position i+1. Taken together, our biochemical data indicate that the major contribution to EBOV RNA synthesis inhibition by remdesivir can be ascribed to delayed chain termination. The long distance of five residues between the incorporated nucleotide analogue and its inhibitory effect warrant further investigation.
Keywords: Ebola virus; GS-5734; RNA polymerase; RdRp; delayed chain termination; remdesivir; respiratory syncytial virus.
Conflict of interest statement
E.P.T. and M.G. declare no conflict of interest. J.Y.F. and D.P.P. are employees of Gilead Sciences, Inc.
Figures


References
-
- WHO List of Blueprint Priority Diseases. [(accessed on 12 March 2019)]; Available online: http://www.who.int/blueprint/priority-diseases/en/
-
- CDC 2014–2016 Ebola Outbreak in West Africa. [(accessed on 12 March 2019)]; Available online: https://www.cdc.gov/vhf/ebola/history/2014-2016-outbreak/index.html.
-
- Henao-Restrepo A.M., Longini I.M., Egger M., Dean N.E., Edmunds W.J., Camacho A., Carroll M.W., Doumbia M., Draguez B., Duraffour S., et al. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet. 2015;386:857–866. doi: 10.1016/S0140-6736(15)61117-5. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

