FGMP: assessing fungal genome completeness
- PMID: 30987585
- PMCID: PMC6466665
- DOI: 10.1186/s12859-019-2782-9
FGMP: assessing fungal genome completeness
Abstract
Background: Inexpensive high-throughput DNA sequencing has democratized access to genetic information for most organisms so that research utilizing a genome or transcriptome of an organism is not limited to model systems. However, the quality of the assemblies of sampled genomes can vary greatly which hampers utility for comparisons and meaningful interpretation. The uncertainty of the completeness of a given genome sequence can limit feasibility of asserting patterns of high rates of gene loss reported in many lineages.
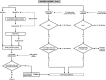
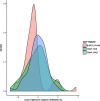
Results: We propose a computational framework and sequence resource for assessing completeness of fungal genomes called FGMP (Fungal Genome Mapping Project). Our approach is based on evolutionary conserved sets of proteins and DNA elements and is applicable to various types of genomic data. We present a comparison of FGMP and state-of-the-art methods for genome completeness assessment utilizing 246 genome assemblies of fungi. We discuss genome assembly improvements/degradations in 57 cases where assemblies have been updated, as recorded by NCBI assembly archive.
Conclusion: FGMP is an accurate tool for quantifying level of completion from fungal genomic data. It is particularly useful for non-model organisms without reference genomes and can be used directly on unassembled reads, which can help reducing genome sequencing costs.
Keywords: Assembly; Conserved elements; Gene model.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Sohn JI, Nam JW. The present and future of de novo whole-genome assembly. Brief Bioinform. 2018;19:23–40. - PubMed
-
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. - DOI - PubMed
-
- Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stuber K, van Themaat EV, Brown JK, Butcher SA, Gurr SJ, Lebrun MH, Ridout CJ, Schulze-Lefert P, Talbot NJ, Ahmadinejad N, Ametz C, Barton GR, Benjdia M, Bidzinski P, Bindschedler LV, Both M, Brewer MT, Cadle-Davidson L, Cadle-Davidson MM, Collemare J, Cramer R, Frenkel O, Godfrey D, Harriman J, Hoede C, King BC, Klages S, Kleemann J, Knoll D, Koti PS, Kreplak J, Lopez-Ruiz FJ, Lu X, Maekawa T, Mahanil S, Micali C, Milgroom MG, Montana G, Noir S, O'Connell RJ, Oberhaensli S, Parlange F, Pedersen C, Quesneville H, Reinhardt R, Rott M, Sacristan S, Schmidt SM, Schon M, Skamnioti P, Sommer H, Stephens A, Takahara H, Thordal-Christensen H, Vigouroux M, Wessling R, Wicker T, Panstruga R. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330:1543–1546. doi: 10.1126/science.1194573. - DOI - PubMed
-
- Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canback B, Choi C, Cichocki N, Clum A, Colpaert J, Copeland A, Costa MD, Dore J, Floudas D, Gay G, Girlanda M, Henrissat B, Herrmann S, Hess J, Hogberg N, Johansson T, Khouja HR, LaButti K, Lahrmann U, Levasseur A, Lindquist EA, Lipzen A, Marmeisse R, Martino E, Murat C, Ngan CY, Nehls U, Plett JM, Pringle A, Ohm RA, Perotto S, Peter M, Riley R, Rineau F, Ruytinx J, Salamov A, Shah F, Sun H, Tarkka M, Tritt A, Veneault-Fourrey C, Zuccaro A, Mycorrhizal Genomics Initiative C, Tunlid A, Grigoriev IV, Hibbett DS, Martin F. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 2015;47:410–415. doi: 10.1038/ng.3223. - DOI - PubMed

