CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation
- PMID: 30988284
- PMCID: PMC6465371
- DOI: 10.1038/s41467-019-09703-y
CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation
Abstract
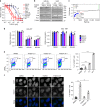
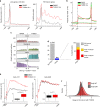
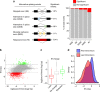
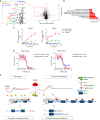
Cyclin-dependent kinase 12 (CDK12) modulates transcription elongation by phosphorylating the carboxy-terminal domain of RNA polymerase II and selectively affects the expression of genes involved in the DNA damage response (DDR) and mRNA processing. Yet, the mechanisms underlying such selectivity remain unclear. Here we show that CDK12 inhibition in cancer cells lacking CDK12 mutations results in gene length-dependent elongation defects, inducing premature cleavage and polyadenylation (PCPA) and loss of expression of long (>45 kb) genes, a substantial proportion of which participate in the DDR. This early termination phenotype correlates with an increased number of intronic polyadenylation sites, a feature especially prominent among DDR genes. Phosphoproteomic analysis indicated that CDK12 directly phosphorylates pre-mRNA processing factors, including those regulating PCPA. These results support a model in which DDR genes are uniquely susceptible to CDK12 inhibition primarily due to their relatively longer lengths and lower ratios of U1 snRNP binding to intronic polyadenylation sites.
Conflict of interest statement
N.S.G. is a founder and equity holder of Syros Pharmaceuticals, C4 Therapeutics, Petra Pharma, Gatekeeper Pharmaceuticals and Soltego. R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics and Dewpoint Therapeutics. R.E.G. is on the SAB of Global Gene Corp. The remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

