Genome analysis of Paenibacillus polymyxa A18 gives insights into the features associated with its adaptation to the termite gut environment
- PMID: 30988376
- PMCID: PMC6465253
- DOI: 10.1038/s41598-019-42572-5
Genome analysis of Paenibacillus polymyxa A18 gives insights into the features associated with its adaptation to the termite gut environment
Abstract
Paenibacillus polymyxa A18 was isolated from termite gut and was identified as a potential cellulase and hemicellulase producer in our previous study. Considering that members belonging to genus Paenibacillus are mostly free-living in soil, we investigated here the essential genetic features that helped P. polymyxa A18 to survive in gut environment. Genome sequencing and analysis identified 4608 coding sequences along with several elements of horizontal gene transfer, insertion sequences, transposases and integrated phages, which add to its genetic diversity. Many genes coding for carbohydrate-active enzymes, including the enzymes responsible for woody biomass hydrolysis in termite gut, were identified in P. polymyxa A18 genome. Further, a series of proteins conferring resistance to 11 antibiotics and responsible for production of 4 antibiotics were also found to be encoded, indicating selective advantage for growth and colonization in the gut environment. To further identify genomic regions unique to this strain, a BLAST-based comparative analysis with the sequenced genomes of 47 members belonging to genus Paenibacillus was carried out. Unique regions coding for nucleic acid modifying enzymes like CRISPR/Cas and Type I Restriction-Modification enzymes were identified in P. polymyxa A18 genome suggesting the presence of defense mechanism to combat viral infections in the gut. In addition, genes responsible for the formation of biofilms, such as Type IV pili and adhesins, which might be assisting P. polymyxa A18 in colonizing the gut were also identified in its genome. In situ colonization experiment further confirmed the ability of P. polymyxa A18 to colonize the gut of termite.
Conflict of interest statement
The authors declare no competing interests.
Figures
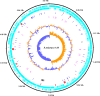


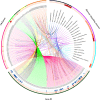
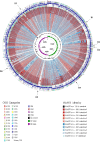

References
-
- Chen, H. In Biotechnology of lignocellulose 25–71 (Springer, 2014).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

