Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase
- PMID: 30988468
- PMCID: PMC6613791
- DOI: 10.1038/s41592-019-0370-6
Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase
Abstract
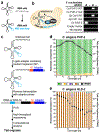
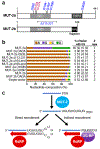
Ribonucleotidyl transferases (rNTases) add untemplated ribonucleotides to diverse RNAs. We have developed TRAID-seq, a screening strategy in Saccharomyces cerevisiae to identify sequences added to a reporter RNA at single-nucleotide resolution by overexpressed candidate enzymes from different organisms. The rNTase activities of 22 previously unexplored enzymes were determined. In addition to poly(A)- and poly(U)-adding enzymes, we identified a cytidine-adding enzyme that is likely to be part of a two-enzyme system that adds CCA to tRNAs in a eukaryote; a nucleotidyl transferase that adds nucleotides to RNA without apparent nucleotide preference; and a poly(UG) polymerase, Caenorhabditis elegans MUT-2, that adds alternating uridine and guanosine nucleotides to form poly(UG) tails. MUT-2 is known to be required for certain forms of RNA silencing, and mutants of the enzyme that result in defective silencing did not add poly(UG) tails in our assay. We propose that MUT-2 poly(UG) polymerase activity is required to promote genome integrity and RNA silencing.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing interests.
Figures



References
METHODS-ONLY REFERENCES
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

