Analysis of oral bacterial communities: comparison of HOMI NGS with a tree-based approach implemented in QIIME
- PMID: 30988892
- PMCID: PMC6450576
- DOI: 10.1080/20002297.2019.1586413
Analysis of oral bacterial communities: comparison of HOMI NGS with a tree-based approach implemented in QIIME
Abstract
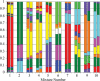
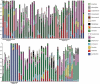
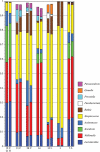
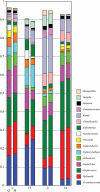
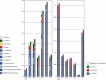
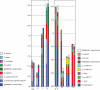
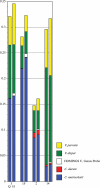
Background: Molecular taxonomic assignments in oral microbial communities have been made using probe-matching approaches, but never compared to those obtained by more readily accepted tree-based approaches. Objective: To compare community composition profiles obtained from a probe-matching approach (HOMINGS) to those from a closed-ended tree-based approach (QIIME using the eHOMD database). Design: HOMINGS and QIIME were used for parallel analysis of ten mock community samples, and of 119 supragingival plaque samples from ecologically unique sites (sound tooth surfaces in healthy subjects, sound tooth surfaces in patients with primary Sjögren's Syndrome, and carious lesions in Sjögren's Syndrome patients). Linear discriminant analysis Effective Size (LEfSe) was used to identify discriminating taxa among the natural plaque samples. Results: Community composition profiles of all samples were congruent between the two analysis aproaches. Alpha and beta diversity of the natural plaque communities were likewise similar. Communities from pSS patients and those from individuals with normal salivary flow differed in alpha and beta diversity. Both classification approaches yielded differences in composition predicted for samples from these subject cohorts, and discriminating taxa were similar between approaches. Conclusions: A direct comparison demonstrates that HOMINGS is largely equivalent to the tree-based approach as implemented here.
Keywords: HOMINGS; QIIME; microbiome; primary Sjögren’s Syndrome; supragingival dental plaque.
Figures

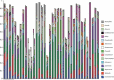

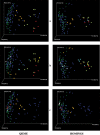
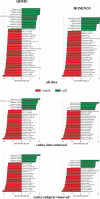
References
-
- Marsh PD, Moter A, Devine DA. Dental plaque biofilms: communities, conflict and control. Periodontol 2000. 2011;55:16–15. PubMed PMID: WOS:000285065800002; English. - PubMed
-
- Socransky SS, Haffajee AD, Cugini MA, et al.. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25(2):134–144. - PubMed
-
- Leung KCM, Leung WK, McMillan AS. Supra-gingival microbiota in Sjögren’s syndrome. Clin Oral Investig. 2007December;11(4):415–423. PubMed PMID: WOS:000251009800014; English. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
