Principles of genome folding into topologically associating domains
- PMID: 30989119
- PMCID: PMC6457944
- DOI: 10.1126/sciadv.aaw1668
Principles of genome folding into topologically associating domains
Abstract
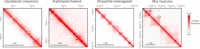
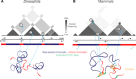
Understanding the mechanisms that underlie chromosome folding within cell nuclei is essential to determine the relationship between genome structure and function. The recent application of "chromosome conformation capture" techniques has revealed that the genome of many species is organized into domains of preferential internal chromatin interactions called "topologically associating domains" (TADs). This chromosome chromosome folding has emerged as a key feature of higher-order genome organization and function through evolution. Although TADs have now been described in a wide range of organisms, they appear to have specific characteristics in terms of size, structure, and proteins involved in their formation. Here, we depict the main features of these domains across species and discuss the relation between chromatin structure, genome activity, and epigenome, highlighting mechanistic principles of TAD formation. We also consider the potential influence of TADs in genome evolution.
Figures

References
-
- Bickmore W. A., The spatial organization of the human genome. Annu. Rev. Genomics Hum. Genet. 14, 67–84 (2013). - PubMed
-
- Bonev B., Cavalli G., Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678 (2016). - PubMed
-
- Dekker J., Rippe K., Dekker M., Kleckner N., Capturing chromosome conformation. Science 295, 1306–1311 (2002). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

