Enantiospecific Pharmacogenomics of Fluvastatin
- PMID: 30989645
- PMCID: PMC6767327
- DOI: 10.1002/cpt.1463
Enantiospecific Pharmacogenomics of Fluvastatin
Abstract
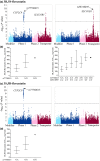
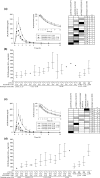
The aim of this study was to investigate how variability in multiple genes related to pharmacokinetics affects fluvastatin exposure. We determined fluvastatin enantiomer pharmacokinetics and sequenced 379 pharmacokinetic genes in 200 healthy volunteers. CYP2C9*3 associated with significantly increased area under the plasma concentration-time curve (AUC) of both 3R,5S-fluvastatin and 3S,5R-fluvastatin (by 67% and 94% per variant allele copy, P = 3.77 × 10-9 and P = 3.19 × 10-12 ). In contrast, SLCO1B1 c.521T>C associated with increased AUC of active 3R,5S-fluvastatin only (by 34% per variant allele copy; P = 8.15 × 10-8 ). A candidate gene analysis suggested that CYP2C9*2 also affects the AUC of both fluvastatin enantiomers and that SLCO2B1 single-nucleotide variations may affect the AUC of 3S,5R-fluvastatin. Thus, SLCO transporters have enantiospecific effects on fluvastatin pharmacokinetics in humans. Genotyping of both CYP2C9 and SLCO1B1 may be useful in predicting fluvastatin efficacy and myotoxicity.
© 2019 The Authors Clinical Pharmacology & Therapeutics published by Wiley Periodicals, Inc. on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
The authors declared no competing interests for this work.
Figures


References
-
- Kathawala, F.G. HMG‐CoA reductase inhibitors: an exciting development in the treatment of hyperlipoproteinemia. Med. Res. Rev. 11, 121–146 (1991). - PubMed
-
- Fischer, V. et al The 3‐hydroxy‐3‐methylglutaryl coenzyme A reductase inhibitor fluvastatin: effect on human cytochrome P‐450 and implications for metabolic drug interactions. Drug Metab. Dispos. 27, 410–416 (1999). - PubMed
-
- Neuvonen, P.J. , Niemi, M. & Backman, J.T. Drug interactions with lipid‐lowering drugs: mechanisms and clinical relevance. Clin. Pharmacol. Ther. 80, 565–581 (2006). - PubMed
-
- Kopplow, K. , Letschert, K. , König, J. , Walter, B. & Keppler, D. Human hepatobiliary transport of organic anions analyzed by quadruple‐transfected cells. Mol. Pharmacol. 68, 1031–1038 (2005). - PubMed
-
- Hirano, M. , Maeda, K. , Matsushima, S. , Nozaki, Y. , Kusuhara, H. & Sugiyama, Y. Involvement of BCRP (ABCG2) in the biliary excretion of pitavastatin. Mol. Pharmacol. 68, 800–807 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

