The 2019 mathematical oncology roadmap
- PMID: 30991381
- PMCID: PMC6655440
- DOI: 10.1088/1478-3975/ab1a09
The 2019 mathematical oncology roadmap
Abstract
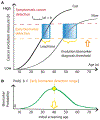
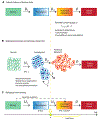
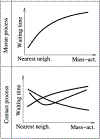
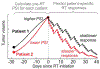
Whether the nom de guerre is Mathematical Oncology, Computational or Systems Biology, Theoretical Biology, Evolutionary Oncology, Bioinformatics, or simply Basic Science, there is no denying that mathematics continues to play an increasingly prominent role in cancer research. Mathematical Oncology-defined here simply as the use of mathematics in cancer research-complements and overlaps with a number of other fields that rely on mathematics as a core methodology. As a result, Mathematical Oncology has a broad scope, ranging from theoretical studies to clinical trials designed with mathematical models. This Roadmap differentiates Mathematical Oncology from related fields and demonstrates specific areas of focus within this unique field of research. The dominant theme of this Roadmap is the personalization of medicine through mathematics, modelling, and simulation. This is achieved through the use of patient-specific clinical data to: develop individualized screening strategies to detect cancer earlier; make predictions of response to therapy; design adaptive, patient-specific treatment plans to overcome therapy resistance; and establish domain-specific standards to share model predictions and to make models and simulations reproducible. The cover art for this Roadmap was chosen as an apt metaphor for the beautiful, strange, and evolving relationship between mathematics and cancer.
Figures








References
Publication types
MeSH terms
Grants and funding
- U01 CA142565/CA/NCI NIH HHS/United States
- R01 CA185486/CA/NCI NIH HHS/United States
- R01 CA164371/CA/NCI NIH HHS/United States
- R13 FD005501/FD/FDA HHS/United States
- K12 CA076917/CA/NCI NIH HHS/United States
- M01 RR000750/RR/NCRR NIH HHS/United States
- MR/P00122X/1/MRC_/Medical Research Council/United Kingdom
- MR/S003851/1/MRC_/Medical Research Council/United Kingdom
- R13 FD005926/FD/FDA HHS/United States
- U54 CA209971/CA/NCI NIH HHS/United States
- U54 CA193313/CA/NCI NIH HHS/United States
- U01 CA232382/CA/NCI NIH HHS/United States
- R01 CA186193/CA/NCI NIH HHS/United States
- U01 CA174706/CA/NCI NIH HHS/United States
- U01 GM111243/GM/NIGMS NIH HHS/United States
- R01 CA179044/CA/NCI NIH HHS/United States
- R13 FD005161/FD/FDA HHS/United States
- R01 GM122424/GM/NIGMS NIH HHS/United States
- R01 NS060752/NS/NINDS NIH HHS/United States
- U54 CA193489/CA/NCI NIH HHS/United States
- OT2 OD026671/OD/NIH HHS/United States
- U01 CA220378/CA/NCI NIH HHS/United States
- U54 CA143970/CA/NCI NIH HHS/United States
- R01 CA100908/CA/NCI NIH HHS/United States
- U54 CA210180/CA/NCI NIH HHS/United States
- U01 CA232137/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
