Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma
- PMID: 30992025
- PMCID: PMC6469146
- DOI: 10.1186/s13046-019-1139-6
Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma
Abstract
Background: Acquired drug resistance is a constraining factor in clinical treatment of glioblastoma (GBM). However, the mechanisms of chemoresponsive tumors acquire therapeutic resistance remain poorly understood. Here, we aim to investigate whether temozolomide (TMZ) resistance of chemoresponsive GBM was enhanced by long non-coding RNA SBF2 antisense RNA 1 (lncRNA SBF2-AS1) enriched exosomes.
Method: LncSBF2-AS1 level in TMZ-resistance or TMZ-sensitive GBM tissues and cells were analyzed by qRT-PCR and FISH assays. A series of in vitro assay and xenograft tumor models were performed to observe the effect of lncSBF2-AS1 on TMZ-resistance in GBM. CHIP assay were used to investigate the correlation of SBF2-AS1 and transcription factor zinc finger E-box binding homeobox 1 (ZEB1). Dual-luciferase reporter, RNA immunoprecipitation (RIP), immunofluorescence and western blotting were performed to verify the relation between lncSBF2-AS1, miR-151a-3p and XRCC4. Comet assay and immunoblotting were performed to expound the effect of lncSBF2-AS1 on DNA double-stand break (DSB) repair. A series of in vitro assay and intracranial xenografts tumor model were used to determined the function of exosomal lncSBF2-AS1.
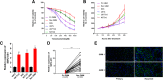
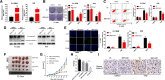
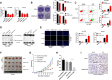
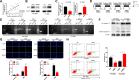
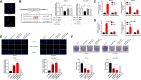
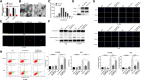
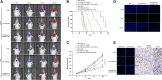
Result: It was found that SBF2-AS1 was upregulated in TMZ-resistant GBM cells and tissues, and overexpression of SBF2-AS1 led to the promotion of TMZ resistance, whereas its inhibition sensitized resistant GBM cells to TMZ. Transcription factor ZEB1 was found to directly bind to the SBF2-AS1 promoter region to regulate SBF2-AS1 level and affected TMZ resistance in GBM cells. SBF2-AS1 functions as a ceRNA for miR-151a-3p, leading to the disinhibition of its endogenous target, X-ray repair cross complementing 4 (XRCC4), which enhances DSB repair in GBM cells. Exosomes selected from temozolomide-resistant GBM cells had high levels of SBF2-AS1 and spread TMZ resistance to chemoresponsive GBM cells. Clinically, high levels of lncSBF2-AS1 in serum exosomes were associated with poor response to TMZ treatment in GBM patients.
Conclusion: We can conclude that GBM cells remodel the tumor microenvironment to promote tumor chemotherapy-resistance by secreting the oncogenic lncSBF2-AS1-enriched exosomes. Thus, exosomal lncSBF2-AS1 in human serum may serve as a possible diagnostic marker for therapy-refractory GBM.
Keywords: Exosomes; Glioblastoma; LncRNA-SBF2-AS1; Temozolomide-resistance.
Conflict of interest statement
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Nanjing Medical University and written informed consent was obtained from all patients.
Consent for publication
We have obtained consents to publish this paper from all the participants.
Competing interests
The authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma.Cancer Lett. 2018 Nov 1;436:10-21. doi: 10.1016/j.canlet.2018.08.004. Epub 2018 Aug 10. Cancer Lett. 2018. PMID: 30102952
-
DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma.Mol Cancer. 2020 Feb 10;19(1):28. doi: 10.1186/s12943-020-1137-5. Mol Cancer. 2020. PMID: 32039732 Free PMC article.
-
Exosomal transfer of miR‑25‑3p promotes the proliferation and temozolomide resistance of glioblastoma cells by targeting FBXW7.Int J Oncol. 2021 Aug;59(2):64. doi: 10.3892/ijo.2021.5244. Epub 2021 Jul 19. Int J Oncol. 2021. PMID: 34278448 Free PMC article.
-
MicroRNAs as the pivotal regulators of Temozolomide resistance in glioblastoma.Mol Brain. 2024 Jul 2;17(1):42. doi: 10.1186/s13041-024-01113-6. Mol Brain. 2024. PMID: 38956588 Free PMC article. Review.
-
Exosomal noncoding RNAs: key players in glioblastoma drug resistance.Mol Cell Biochem. 2021 Nov;476(11):4081-4092. doi: 10.1007/s11010-021-04221-2. Epub 2021 Jul 17. Mol Cell Biochem. 2021. PMID: 34273059 Review.
Cited by
-
LncRNA HAS2-AS1 Promotes Glioblastoma Proliferation by Sponging miR-137.Front Oncol. 2021 May 20;11:634893. doi: 10.3389/fonc.2021.634893. eCollection 2021. Front Oncol. 2021. PMID: 34094916 Free PMC article.
-
The Role of Intracellular and Extracellular Vesicles in the Development of Therapy Resistance in Cancer.Curr Pharm Des. 2024;30(35):2765-2784. doi: 10.2174/0113816128326325240723051625. Curr Pharm Des. 2024. PMID: 39113303 Review.
-
Evaluation of the genetic basis of familial-associated early-onset hematologic cancers in an ancestral/ethnically diverse population.Haematologica. 2024 Jul 1;109(7):2085-2091. doi: 10.3324/haematol.2023.284224. Haematologica. 2024. PMID: 38205536 Free PMC article.
-
Extracellular Vesicles: Emerging Modulators of Cancer Drug Resistance.Cancers (Basel). 2021 Feb 11;13(4):749. doi: 10.3390/cancers13040749. Cancers (Basel). 2021. PMID: 33670185 Free PMC article. Review.
-
Drug Resistance in Cancers: A Free Pass for Bullying.Cells. 2022 Oct 26;11(21):3383. doi: 10.3390/cells11213383. Cells. 2022. PMID: 36359776 Free PMC article. Review.
References
-
- Stupp R, Hegi M, Mason W, van den Bent M, Taphoorn M, Janzer R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

