Single-cell whole-genome sequencing reveals the functional landscape of somatic mutations in B lymphocytes across the human lifespan
- PMID: 30992375
- PMCID: PMC6500118
- DOI: 10.1073/pnas.1902510116
Single-cell whole-genome sequencing reveals the functional landscape of somatic mutations in B lymphocytes across the human lifespan
Abstract
Accumulation of mutations in somatic cells has been implicated as a cause of aging since the 1950s. However, attempts to establish a causal relationship between somatic mutations and aging have been constrained by the lack of methods to directly identify mutational events in primary human tissues. Here we provide genome-wide mutation frequencies and spectra of human B lymphocytes from healthy individuals across the entire human lifespan using a highly accurate single-cell whole-genome sequencing method. We found that the number of somatic mutations increases from <500 per cell in newborns to >3,000 per cell in centenarians. We discovered mutational hotspot regions, some of which, as expected, were located at Ig genes associated with somatic hypermutation (SHM). B cell-specific mutation signatures associated with development, aging, or SHM were found. The SHM signature strongly correlated with the signature found in human B cell tumors, indicating that potential cancer-causing events are already present even in B cells of healthy individuals. We also identified multiple mutations in sequence features relevant to cellular function (i.e., transcribed genes and gene regulatory regions). Such mutations increased significantly during aging, but only at approximately one-half the rate of the genome average, indicating selection against mutations that impact B cell function. This full characterization of the landscape of somatic mutations in human B lymphocytes indicates that spontaneous somatic mutations accumulating with age can be deleterious and may contribute to both the increased risk for leukemia and the functional decline of B lymphocytes in the elderly.
Keywords: B lymphocyte; aging; functional genome; single-cell whole-genome sequencing; somatic DNA mutation.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: L.Z., X.D., M.L., A.Y.M., and J.V. are cofounders of SingulOmics Corp.
Figures

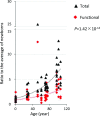
References
-
- Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

