The BIN1 rs744373 SNP is associated with increased tau-PET levels and impaired memory
- PMID: 30992433
- PMCID: PMC6467911
- DOI: 10.1038/s41467-019-09564-5
The BIN1 rs744373 SNP is associated with increased tau-PET levels and impaired memory
Abstract
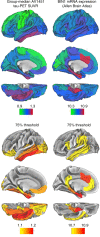
The single nucleotide polymorphism (SNP) rs744373 in the bridging integrator-1 gene (BIN1) is a risk factor for Alzheimer's disease (AD). In the brain, BIN1 is involved in endocytosis and sustaining cytoskeleton integrity. Post-mortem and in vitro studies suggest that BIN1-associated AD risk is mediated by increased tau pathology but whether rs744373 is associated with increased tau pathology in vivo is unknown. Here we find in 89 older individuals without dementia, that BIN1 rs744373 risk-allele carriers show higher AV1451 tau-PET across brain regions corresponding to Braak stages II-VI. In contrast, the BIN1 rs744373 SNP was not associated with AV45 amyloid-PET uptake. Furthermore, the rs744373 risk-allele was associated with worse memory performance, mediated by increased global tau levels. Together, our findings suggest that the BIN1 rs744373 SNP is associated with increased tau but not beta-amyloid pathology, suggesting that alterations in BIN1 may contribute to memory deficits via increased tau pathology.
Conflict of interest statement
The authors declare no competing interests.
Figures