Bioproduction of pure, kilobase-scale single-stranded DNA
- PMID: 30992517
- PMCID: PMC6467869
- DOI: 10.1038/s41598-019-42665-1
Bioproduction of pure, kilobase-scale single-stranded DNA
Abstract
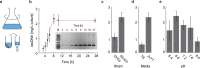
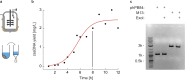
Scalable production of kilobase single-stranded DNA (ssDNA) with sequence control has applications in therapeutics, gene synthesis and sequencing, scaffolded DNA origami, and archival DNA memory storage. Biological production of circular ssDNA (cssDNA) using M13 addresses these needs at low cost. However, one unmet goal is to minimize the essential protein coding regions of the exported DNA while maintaining its infectivity and production purity to produce sequences less than 3,000 nt in length, relevant to therapeutic and materials science applications. Toward this end, synthetic miniphage with inserts of custom sequence and size offers scalable, low-cost synthesis of cssDNA at milligram and higher scales. Here, we optimize growth conditions using an E. coli helper strain combined with a miniphage genome carrying only an f1 origin and a β-lactamase-encoding (bla) antibiotic resistance gene, enabling isolation of pure cssDNA with a minimum sequence genomic length of 1,676 nt, without requiring additional purification from contaminating DNA. Low-cost scalability of isogenic, custom-length cssDNA is demonstrated for a sequence of 2,520 nt using a bioreactor, purified with low endotoxin levels (<5 E.U./ml). We apply these exonuclease-resistant cssDNAs to the self-assembly of wireframe DNA origami objects and to encode digital information on the miniphage genome for biological amplification.
Conflict of interest statement
T.R.S., R.R.D. and M.B. are coinventors on a patent pending (62/584,664) for some of the methods disclosed herein.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

