Precise definition of PTEN C-terminal epitopes and its implications in clinical oncology
- PMID: 30993208
- PMCID: PMC6465295
- DOI: 10.1038/s41698-019-0083-4
Precise definition of PTEN C-terminal epitopes and its implications in clinical oncology
Abstract
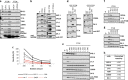
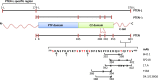
Anti-PTEN monoclonal antibodies (mAb) are arising as important tools for immunohistochemistry (IHC) and protein quantification routine analysis in clinical oncology. Although an effort has been made to document the reliability of tumor tissue section immunostaining by anti-PTEN mAb, and to standardize their IHC use in research and in the clinical practice, the precise topological and biochemical definition of the epitope recognized by each mAb has been conventionally overlooked. In this study, six commercial anti-PTEN mAb have been validated and characterized for sensitivity and specificity by IHC and FISH, using a set of prostate and urothelial bladder tumor specimens, and by immunoblot, using PTEN positive and PTEN negative human cell lines. Immunoblot precise epitope mapping, performed using recombinant PTEN variants and mutations, revealed that all mAb recognized linear epitopes of 6-11 amino acid length at the PTEN C-terminus. Tumor-associated or disease-associated mutations at the PTEN C-terminus did not affect subcellular localization or PIP3 phosphatase activity of PTEN in cells, although resulted in specific loss of reactivity for some mAb. Furthermore, specific mimicking-phosphorylation mutations at the PTEN C-terminal region also abolished binding of specific mAb. Our study adds new evidence on the relevance of a precise epitope mapping in the validation of anti-PTEN mAb for their use in the clinics. This will be substantial to provide a more accurate diagnosis in clinical oncology based on PTEN protein expression in tumors and biological fluids.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

