Whole-genome Sequencing Provides Data for Stratifying Infection Prevention and Control Management of Nosocomial Influenza A
- PMID: 30993315
- PMCID: PMC6821348
- DOI: 10.1093/cid/ciz020
Whole-genome Sequencing Provides Data for Stratifying Infection Prevention and Control Management of Nosocomial Influenza A
Abstract
Background: Influenza A virus causes annual epidemics in humans and is associated with significant morbidity and mortality. Haemagglutinin (HA) and neuraminidase (NA) gene sequencing have traditionally been used to identify the virus genotype, although their utility in detecting outbreak clusters is still unclear. The objective of this study was to determine the utility, if any, of whole-genome sequencing over HA/NA sequencing for infection prevention and control (IPC) in hospitals.
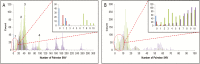
Methods: We obtained all clinical samples from influenza (H1N1)-positive patients at the Great Ormond Street Hospital between January and March 2016. Samples were sequenced using targeted enrichment on an Illumina MiSeq sequencer. Maximum likelihood trees were computed for both whole genomes and concatenated HA/NA sequences. Epidemiological data was taken from routine IPC team activity during the period.
Results: Complete genomes were obtained for 65/80 samples from 38 patients. Conventional IPC analysis recognized 1 outbreak, involving 3 children, and identified another potential cluster in the haemato-oncology ward. Whole-genome and HA/NA phylogeny both accurately identified the previously known outbreak cluster. However, HA/NA sequencing additionally identified unrelated strains as part of this outbreak cluster. A whole-genome analysis identified a further cluster of 2 infections that had been previously missed and refuted suspicions of transmission in the haemato-oncology wards.
Conclusions: Whole-genome sequencing is better at identifying outbreak clusters in a hospital setting than HA/NA sequencing. Whole-genome sequencing could provide a faster and more reliable method for outbreak monitoring and supplement routine IPC team work to allow the prevention of transmission.
Keywords: infection control; influenza; next-generation sequencing; transmission; whole genome.
© The Author(s) 2019. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures



References
-
- Mauskopf J, Klesse M, Lee S, Herrera-Taracena G. The burden of influenza complications in different high-risk groups: a targeted literature review. J Med Econ 2013; 16:264–77. - PubMed
-
- Grund S, Roggendorf M, Schweiger B. Outbreak of influenza virus A/H1N1 in a hospital ward for immunocompromised patients. Arch Virol 2010; 155:1797–802. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

