Neuronal Activity in Non-LNv Clock Cells Is Required to Produce Free-Running Rest:Activity Rhythms in Drosophila
- PMID: 30994046
- PMCID: PMC7153773
- DOI: 10.1177/0748730419841468
Neuronal Activity in Non-LNv Clock Cells Is Required to Produce Free-Running Rest:Activity Rhythms in Drosophila
Abstract
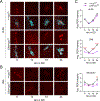
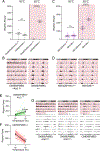
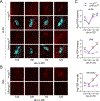
Circadian rhythms in behavior and physiology are produced by central brain clock neurons that can be divided into subpopulations based on molecular and functional characteristics. It has become clear that coherent behavioral rhythms result from the coordinated action of these clock neuron populations, but many questions remain regarding the organizational logic of the clock network. Here we used targeted genetic tools in Drosophila to eliminate either molecular clock function or neuronal activity in discrete clock neuron subsets. We find that neuronal firing is necessary across multiple clock cell populations to produce free-running rhythms of rest and activity. In contrast, such rhythms are much more subtly affected by molecular clock suppression in the same cells. These findings demonstrate that network connectivity can compensate for a lack of molecular oscillations within subsets of clock cells. We further show that small ventrolateral (sLNv) clock neurons, which have been characterized as master pacemakers under free-running conditions, cannot drive rhythms independent of communication between other cells of the clock network. In particular, we pinpoint an essential contribution of the dorsolateral (LNd) clock neurons, and show that manipulations that affect LNd function reduce circadian rhythm strength without affecting molecular cycling in sLNv cells. These results suggest a hierarchical organization in which circadian information is first consolidated among one or more clock cell populations before accessing output pathways that control locomotor activity.
Keywords: LNd; circadian; circuit; sLNv.
Conflict of interest statement
Figures




References
-
- Abruzzi KC, Zadina A, Luo W, Wiyanto E, Rahman R, Guo F, Shafer O, Rosbash M (2017) RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLOS Genet 13:e1006613 Available at: http://dx.plos.org/10.1371/journal.pgen.1006613. - DOI - PMC - PubMed
-
- Beckwith EJ, Ceriani MF (2015) Experimental assessment of the network properties of the Drosophila circadian clock. J Comp Neurol 523:982–996. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

