A computational solution to improve biomarker reproducibility during long-term projects
- PMID: 30995241
- PMCID: PMC6469750
- DOI: 10.1371/journal.pone.0209060
A computational solution to improve biomarker reproducibility during long-term projects
Abstract
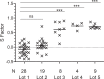
Biomarkers are fundamental to basic and clinical research outcomes by reporting host responses and providing insight into disease pathophysiology. Measuring biomarkers with research-use ELISA kits is universal, yet lack of kit standardization and unexpected lot-to-lot variability presents analytic challenges for long-term projects. During an ongoing two-year project measuring plasma biomarkers in cancer patients, control concentrations for one biomarker (PF) decreased significantly after changes in ELISA kit lots. A comprehensive operations review pointed to standard curve shifts with the new kits, an analytic variable that jeopardized data already collected on hundreds of patient samples. After excluding other reasonable contributors to data variability, a computational solution was developed to provide a uniform platform for data analysis across multiple ELISA kit lots. The solution (ELISAtools) was developed within open-access R software in which variability between kits is treated as a batch effect. A defined best-fit Reference standard curve is modelled, a unique Shift factor "S" is calculated for every standard curve and data adjusted accordingly. The averaged S factors for PF ELISA kit lots #1-5 ranged from -0.086 to 0.735, and reduced control inter-assay variability from 62.4% to <9%, within quality control limits. S factors calculated for four other biomarkers provided a quantitative metric to monitor ELISAs over the 10 month study period for quality control purposes. Reproducible biomarker measurements are essential, particularly for long-term projects with valuable patient samples. Use of research-use ELISA kits is ubiquitous and judicious use of this computational solution maximizes biomarker reproducibility.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

