Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated?
- PMID: 30995449
- PMCID: PMC6544567
- DOI: 10.1016/j.jmb.2019.04.009
Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated?
Abstract
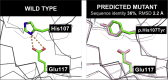
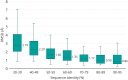
Knowledge of protein structure can be used to predict the phenotypic consequence of a missense variant. Since structural coverage of the human proteome can be roughly tripled to over 50% of the residues if homology-predicted structures are included in addition to experimentally determined coordinates, it is important to assess the reliability of using predicted models when analyzing missense variants. Accordingly, we assess whether a missense variant is structurally damaging by using experimental and predicted structures. We considered 606 experimental structures and show that 40% of the 1965 disease-associated missense variants analyzed have a structurally damaging change in the mutant structure. Only 11% of the 2134 neutral variants are structurally damaging. Importantly, similar results are obtained when 1052 structures predicted using Phyre2 algorithm were used, even when the model shares low (<40%) sequence identity to the template. Thus, structure-based analysis of the effects of missense variants can be effectively applied to homology models. Our in-house pipeline, Missense3D, for structurally assessing missense variants was made available at http://www.sbg.bio.ic.ac.uk/~missense3d.
Keywords: Phyre2 protein structure prediction; missense variants; protein structure prediction; structure-based prediction; variant effect prediction.
Copyright © 2019 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures






References
-
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. - PMC - PubMed
-
- Ellard S, Baple EL, Owens M, Cannon S, Eccles DM, Abbs S, et al. ACGS best practice guidelines for variant classification 2017. ACGS 2017:1–16.

