Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors
- PMID: 30995674
- PMCID: PMC6657343
- DOI: 10.1038/s41586-019-1161-z
Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors
Abstract
CRISPR-Cas base-editor technology enables targeted nucleotide alterations, and is being increasingly used for research and potential therapeutic applications1,2. The most widely used cytosine base editors (CBEs) induce deamination of DNA cytosines using the rat APOBEC1 enzyme, which is targeted by a linked Cas protein-guide RNA complex3,4. Previous studies of the specificity of CBEs have identified off-target DNA edits in mammalian cells5,6. Here we show that a CBE with rat APOBEC1 can cause extensive transcriptome-wide deamination of RNA cytosines in human cells, inducing tens of thousands of C-to-U edits with frequencies ranging from 0.07% to 100% in 38-58% of expressed genes. CBE-induced RNA edits occur in both protein-coding and non-protein-coding sequences and generate missense, nonsense, splice site, and 5' and 3' untranslated region mutations. We engineered two CBE variants bearing mutations in rat APOBEC1 that substantially decreased the number of RNA edits (by more than 390-fold and more than 3,800-fold) in human cells. These variants also showed more precise on-target DNA editing than the wild-type CBE and, for most guide RNAs tested, no substantial reduction in editing efficiency. Finally, we show that an adenine base editor7 can also induce transcriptome-wide RNA edits. These results have implications for the use of base editors in both research and clinical settings, illustrate the feasibility of engineering improved variants with reduced RNA editing activities, and suggest the need to more fully define and characterize the RNA off-target effects of deaminase enzymes in base editor platforms.
Conflict of interest statement
Competing Interests Statement: J.K.J. has financial interests in Beam Therapeutics, Editas Medicine, Endcadia, Pairwise Plants, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.K.J. is a member of the Board of Directors of the American Society of Gene and Cell Therapy. J.G., R.Z., and J.K.J. are co-inventors on patent applications that have been filed by Partners Healthcare/Massachusetts General Hospital on engineered base editor architectures that reduce RNA editing activities.
Figures





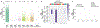


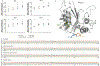

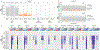

Comment in
-
Towards precise, safe genome editing.Cell Res. 2019 Sep;29(9):687-689. doi: 10.1038/s41422-019-0188-x. Cell Res. 2019. PMID: 31201381 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

