Oligomeric Architecture of Mouse Activating Nkrp1 Receptors on Living Cells
- PMID: 30995786
- PMCID: PMC6515139
- DOI: 10.3390/ijms20081884
Oligomeric Architecture of Mouse Activating Nkrp1 Receptors on Living Cells
Abstract
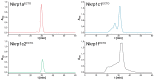
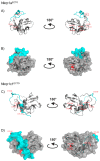
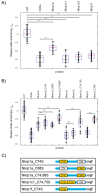
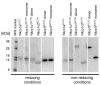
Mouse activating Nkrp1 proteins are commonly described as type II transmembrane receptors with disulfide-linked homodimeric structure. Their function and the manner in which Nkrp1 proteins of mouse strain (C57BL/6) oligomerize are still poorly understood. To assess the oligomerization state of Nkrp1 proteins, mouse activating EGFP-Nkrp1s were expressed in mammalian lymphoid cells and their oligomerization evaluated by Förster resonance energy transfer (FRET). Alternatively, Nkrp1s oligomers were detected by Western blotting to specify the ratio between monomeric and dimeric forms. We also performed structural characterization of recombinant ectodomains of activating Nkrp1 receptors. Nkrp1 isoforms c1, c2 and f were expressed prevalently as homodimers, whereas the Nkrp1a displays larger proportion of monomers on the cell surface. Cysteine-to-serine mutants revealed the importance of all stalk cysteines for protein dimerization in living cells with a major influence of cysteine at position 74 in two Nkrp1 protein isoforms. Our results represent a new insight into the oligomerization of Nkrp1 receptors on lymphoid cells, which will help to determine their function.
Keywords: Förster resonance energy transfer; Nkrp1; cysteine; dimerization; disulfide bond arrangement.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Giorda R., Trucco M. Mouse NKR-P1. A family of genes selectively coexpressed in adherent lymphokine-activated killer cells. J. Immunol. 1991;147:1701–1708. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

