Filamentous bacteriophages are associated with chronic Pseudomonas lung infections and antibiotic resistance in cystic fibrosis
- PMID: 30996083
- PMCID: PMC7021451
- DOI: 10.1126/scitranslmed.aau9748
Filamentous bacteriophages are associated with chronic Pseudomonas lung infections and antibiotic resistance in cystic fibrosis
Abstract
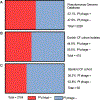
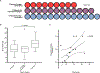
Filamentous bacteriophage (Pf phage) contribute to the virulence of Pseudomonas aeruginosa infections in animal models, but their relevance to human disease is unclear. We sought to interrogate the prevalence and clinical relevance of Pf phage in patients with cystic fibrosis (CF) using sputum samples from two well-characterized patient cohorts. Bacterial genomic analysis in a Danish longitudinal cohort of 34 patients with CF revealed that 26.5% (n = 9) were consistently Pf phage positive. In the second cohort, a prospective cross-sectional cohort of 58 patients with CF at Stanford, sputum qPCR analysis showed that 36.2% (n = 21) of patients were Pf phage positive. In both cohorts, patients positive for Pf phage were older, and in the Stanford CF cohort, patients positive for Pf phage were more likely to have chronic P. aeruginosa infection and had greater declines in pulmonary function during exacerbations than patients negative for Pf phage presence in the sputum. Last, P. aeruginosa strains carrying Pf phage exhibited increased resistance to antipseudomonal antibiotics. Mechanistically, in vitro analysis showed that Pf phage sequesters these same antibiotics, suggesting that this mechanism may thereby contribute to the selection of antibiotic resistance over time. These data provide evidence that Pf phage may contribute to clinical outcomes in P. aeruginosa infection in CF.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures



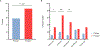

References
-
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iannuzzi MC, Collins FS, Tsui L-C, Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 245, 1066–1073 (1989). - PubMed
-
- Welsh MJ, Ramsey BW, Accurso F, Cutting GR, in The Online Metabolic and Molecular Bases of Inherited Disease, Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, Gibson KM, Mitchell G, Eds. (The McGraw-Hill Companies Inc, 2014).
-
- Khan TZ, Wagener JS, Bost T, Martinez J, Accurso FJ, Riches DW, Early pulmonary inflammation in infants with cystic fibrosis. Am. J. Respir. Crit. Care Med 151, 1075–1082 (1995). - PubMed
-
- Davis PB, Drumm M, Konstan MW, Cystic fibrosis. Am. J. Respir. Crit. Care Med 154, 1229–1256 (1996). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

