Regulation of Human Cytomegalovirus Secondary Envelopment by a C-Terminal Tetralysine Motif in pUL71
- PMID: 30996102
- PMCID: PMC6580969
- DOI: 10.1128/JVI.02244-18
Regulation of Human Cytomegalovirus Secondary Envelopment by a C-Terminal Tetralysine Motif in pUL71
Abstract
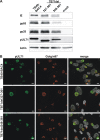
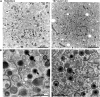
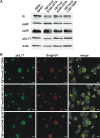
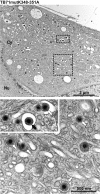
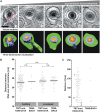
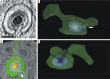
Human cytomegalovirus (HCMV) secondary envelopment requires the viral tegument protein pUL71. The lack of pUL71 results in a complex ultrastructural phenotype with increased numbers of viral capsids undergoing envelopment at the cytoplasmic virus assembly complex. Here, we report a role of the pUL71 C terminus in secondary envelopment. Mutant viruses expressing C-terminally truncated pUL71 (TB71del327-361 and TB71del348-351) exhibited an impaired secondary envelopment in transmission electron microscopy (TEM) studies. Further mutational analyses of the C terminus revealed a tetralysine motif whose mutation (TB71mutK348-351A) resulted in an envelopment defect that was undistinguishable from the defect caused by truncation of the pUL71 C terminus. Interestingly, not all morphological alterations that define the ultrastructural phenotype of a TB71stop virus were found in cells infected with the C-terminally mutated viruses. This suggests that pUL71 provides additional functions that modulate HCMV morphogenesis and are harbored elsewhere in pUL71. This is also reflected by an intermediate growth defect of the C-terminally mutated viruses compared to the growth of the TB71stop virus. Electron tomography and three-dimensional visualization of different stages of secondary envelopment in TB71mutK348-351A-infected cells showed unambiguously the formation of a bud neck. Furthermore, we provide evidence for progressive tegument formation linked to advancing grades of capsid envelopment, suggesting that tegumentation and envelopment are intertwined processes. Altogether, we identified the importance of the pUL71 C terminus and, specifically, of a positively charged tetralysine motif for HCMV secondary envelopment.IMPORTANCE Human cytomegalovirus (HCMV) is an important human pathogen that causes severe symptoms, especially in immunocompromised hosts. Furthermore, congenital HCMV infection is the leading viral cause of severe birth defects. Development of antiviral drugs to prevent the production of infectious virus progeny is challenging due to a complex and multistep virion morphogenesis. The mechanism of secondary envelopment is still not fully understood; nevertheless, it represents a potential target for antiviral drugs. Our identification of the role of a positively charged motif in the pUL71 C terminus for efficient HCMV secondary envelopment underlines the importance of pUL71 and, especially, its C terminus for this process. It furthermore shows how cell-associated spread and virion release depend on secondary envelopment. Ultrastructural analyses of different stages of envelopment contribute to a better understanding of the mechanisms underlying the process of secondary envelopment. This may bring us closer to the development of novel concepts to treat HCMV infections.
Keywords: HCMV; Human betaherpesvirus 5; TEM analysis; UL71; secondary envelopment; tetralysine motif.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Mocarski E, Shenk T, Pass R. 2006. Cytomegaloviruses, p. 2701–2771. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA (ed), Fields virology 5th ed Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol 78:10960–10966. doi:10.1128/JVI.78.20.10960-10966.2004. - DOI - PMC - PubMed
-
- Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol 74:975–986. doi:10.1128/JVI.74.2.975-986.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

