Prediction of UGT-mediated Metabolism Using the Manually Curated MetaQSAR Database
- PMID: 30996809
- PMCID: PMC6466832
- DOI: 10.1021/acsmedchemlett.8b00603
Prediction of UGT-mediated Metabolism Using the Manually Curated MetaQSAR Database
Abstract
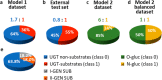
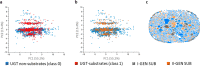
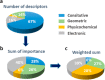
Even though glucuronidations are the most frequent metabolic reactions of conjugation, both in quantitative and qualitative terms, they have rather seldom been investigated using computational approaches. To fill this gap, we have used the manually collected MetaQSAR metabolic reaction database to generate two models for the prediction of UGT-mediated metabolism, both based on molecular descriptors and implementing the Random Forest algorithm. The first model predicts the occurrence of the reaction and was internally validated with a Matthew correlation coefficient (MCC) of 0.76 and an area under the ROC curve (AUC) of 0.94, and further externally validated using a test set composed of 120 additional xenobiotics (MCC of 0.70 and AUC of 0.90). The second model distinguishes between O- and N-glucuronidations and was optimized by the random undersampling procedure to improve the predictive accuracy during the internal validation, with the recall measure of the minority class increasing from 0.55 to 0.78.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

