Single-stranded regions modulate conformational dynamics and ATPase activity of eIF4A to optimize 5'-UTR unwinding
- PMID: 30997503
- PMCID: PMC6547412
- DOI: 10.1093/nar/gkz254
Single-stranded regions modulate conformational dynamics and ATPase activity of eIF4A to optimize 5'-UTR unwinding
Abstract
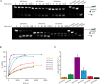
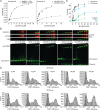
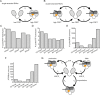
Eukaryotic translation initiation requires unwinding of secondary structures in the 5'-untranslated region of mRNA. The DEAD-box helicase eIF4A is thought to unwind structural elements in the 5'-UTR in conjunction with eIF4G and eIF4B. Both factors jointly stimulate eIF4A activities by modulation of eIF4A conformational cycling between open and closed states. Here we examine how RNA substrates modulate eIF4A activities. The RNAs fall into two classes: Short RNAs only partially stimulate the eIF4A ATPase activity, and closing is rate-limiting for the conformational cycle. By contrast, longer RNAs maximally stimulate ATP hydrolysis and promote closing of eIF4A. Strikingly, the rate constants of unwinding do not correlate with the length of a single-stranded region preceding a duplex, but reach a maximum for RNA with a single-stranded region of six nucleotides. We propose a model in which RNA substrates affect eIF4A activities by modulating the kinetic partitioning of eIF4A between futile, unproductive, and productive cycles.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Grifo J.A., Tahara S.M., Morgan M.A., Shatkin A.J., Merrick W.C.. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 1983; 258:5804–5810. - PubMed
-
- Goyer C., Altmann M., Lee H.S., Blanc A., Deshmukh M., Woolford J.L. Jr, Trachsel H., Sonenberg N.. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell Biol. 1993; 13:4860–4874. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

