Identification of noncoding RNA expression profiles and regulatory interaction networks following traumatic spinal cord injury by sequence analysis
- PMID: 30998503
- PMCID: PMC6520015
- DOI: 10.18632/aging.101919
Identification of noncoding RNA expression profiles and regulatory interaction networks following traumatic spinal cord injury by sequence analysis
Abstract
Aim: To systematically profile and characterize the noncoding RNA (ncRNA) expression pattern in the lesion epicenter of spinal tissues after traumatic spinal cord injury (TSCI) and predicted the structure and potential functions of the regulatory networks associated with these differentially expressed ncRNAs and mRNAs.
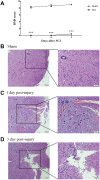
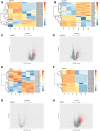
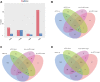
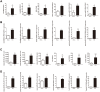
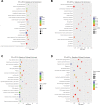
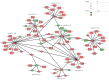
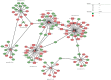
Results: A total of 498 circRNAs, 458 lncRNAs, 155 miRNAs and 1203 mRNAs were identified in TSCI mice models to be differentially expressed. The regulatory networks associated with these differentially expressed ncRNAs and mRNAs were constructed.
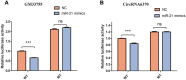
Materials and methods: We used RNA-Seq, Gene ontology (GO), KEGG pathway analysis and co-expression network analyses to profle the expression and regulation patterns of noncoding RNAs and mRNAs of mice models after TSCI. The findings were validated by quantitative real-time PCR (qRT-PCR) and Luciferase assay.
Conclusion: noncoding RNAs might play important roles via the competing endogenous RNA regulation pattern after TSCI, further findings arising from this study will not only expand the understanding of potential ncRNA biomarkers but also help guide therapeutic strategies for TSCI.
Keywords: axonal regeneration; competing endogenous RNA; noncoding RNA; traumatic spinal cord injury.
Conflict of interest statement
Figures
Similar articles
-
Genome-wide analysis of acute traumatic spinal cord injury-related RNA expression profiles and uncovering of a regulatory axis in spinal fibrotic scars.Cell Prolif. 2021 Jan;54(1):e12951. doi: 10.1111/cpr.12951. Epub 2020 Nov 5. Cell Prolif. 2021. PMID: 33150698 Free PMC article.
-
Identification and coregulation pattern analysis of long noncoding RNAs following subacute spinal cord injury.J Orthop Res. 2022 Mar;40(3):661-673. doi: 10.1002/jor.25101. Epub 2021 Jun 6. J Orthop Res. 2022. PMID: 33991009 Free PMC article.
-
[Identification of potential traumatic spinal cord injury related circular RNA-microRNA networks by sequence analysis].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2020 Feb 15;34(2):213-219. doi: 10.7507/1002-1892.201905079. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2020. PMID: 32030954 Free PMC article. Chinese.
-
[Non-coding RNAs as therapeutic targets in spinal cord injury].Zh Vopr Neirokhir Im N N Burdenko. 2020;84(4):104-110. doi: 10.17116/neiro202084031104. Zh Vopr Neirokhir Im N N Burdenko. 2020. PMID: 32759933 Review. Russian.
-
Long non-coding RNAs in the spinal cord injury: Novel spotlight.J Cell Mol Med. 2019 Aug;23(8):4883-4890. doi: 10.1111/jcmm.14422. Epub 2019 May 29. J Cell Mol Med. 2019. PMID: 31140726 Free PMC article. Review.
Cited by
-
miR-182-5p Regulates Nogo-A Expression and Promotes Neurite Outgrowth of Hippocampal Neurons In Vitro.Pharmaceuticals (Basel). 2022 Apr 25;15(5):529. doi: 10.3390/ph15050529. Pharmaceuticals (Basel). 2022. PMID: 35631355 Free PMC article.
-
The emerging role of circular RNAs in spinal cord injury.J Orthop Translat. 2021 Jul 22;30:1-5. doi: 10.1016/j.jot.2021.06.001. eCollection 2021 Sep. J Orthop Translat. 2021. PMID: 34401327 Free PMC article. Review.
-
Circular RNA Plek promotes fibrogenic activation by regulating the miR-135b-5p/TGF-βR1 axis after spinal cord injury.Aging (Albany NY). 2021 May 11;13(9):13211-13224. doi: 10.18632/aging.203002. Epub 2021 May 11. Aging (Albany NY). 2021. PMID: 33982670 Free PMC article.
-
Expression Profile of Long Noncoding RNAs and Circular RNAs in Mouse C3H10T1/2 Mesenchymal Stem Cells Undergoing Myogenic and Cardiomyogenic Differentiation.Stem Cells Int. 2021 Apr 30;2021:8882264. doi: 10.1155/2021/8882264. eCollection 2021. Stem Cells Int. 2021. PMID: 34012468 Free PMC article.
-
Identifying specific miRNAs and associated mRNAs in CD44 and CD90 cancer stem cell subtypes in gastric cancer cell line SNU-5.Int J Clin Exp Pathol. 2020 Jun 1;13(6):1313-1323. eCollection 2020. Int J Clin Exp Pathol. 2020. PMID: 32661467 Free PMC article.
References
-
- van Middendorp JJ, Barbagallo G, Schuetz M, Hosman AJ. Design and rationale of a Prospective, Observational European Multicenter study on the efficacy of acute surgical decompression after traumatic Spinal Cord Injury: the SCI-POEM study Spinal Cord. 2012; 50:686–94. 10.1038/sc.2012.34 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

