Genome-wide association study of endo-parasite phenotypes using imputed whole-genome sequence data in dairy and beef cattle
- PMID: 30999842
- PMCID: PMC6471778
- DOI: 10.1186/s12711-019-0457-7
Genome-wide association study of endo-parasite phenotypes using imputed whole-genome sequence data in dairy and beef cattle
Abstract
Background: Quantitative genetic studies suggest the existence of variation at the genome level that affects the ability of cattle to resist to parasitic diseases. The objective of the current study was to identify regions of the bovine genome that are associated with resistance to endo-parasites.
Methods: Individual cattle records were available for Fasciola hepatica-damaged liver from 18 abattoirs. Deregressed estimated breeding values (EBV) for F. hepatica-damaged liver were generated for genotyped animals with a record for F. hepatica-damaged liver and for genotyped sires with a least one progeny record for F. hepatica-damaged liver; 3702 animals were available. In addition, individual cow records for antibody response to F. hepatica on 6388 genotyped dairy cows, antibody response to Ostertagia ostertagi on 8334 genotyped dairy cows and antibody response to Neospora caninum on 4597 genotyped dairy cows were adjusted for non-genetic effects. Genotypes were imputed to whole-sequence; after edits, 14,190,141 single nucleotide polymorphisms (SNPs) and 16,603,644 SNPs were available for cattle with deregressed EBV for F. hepatica-damaged liver and cows with an antibody response to a parasitic disease, respectively. Association analyses were undertaken using linear regression on one SNP at a time, in which a genomic relationship matrix accounted for the relationships between animals.
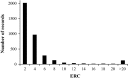
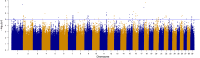
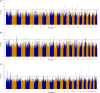
Results: Genomic regions for F. hepatica-damaged liver were located on Bos taurus autosomes (BTA) 1, 8, 11, 16, 17 and 18; each region included at least one SNP with a p value lower than 10-6. Five SNPs were identified as significant (q value < 0.05) for antibody response to N. caninum and were located on BTA21 or 25. For antibody response to F. hepatica and O. ostertagi, six and nine quantitative trait loci (QTL) regions that included at least one SNP with a p value lower than 10-6 were identified, respectively. Gene set enrichment analysis revealed a significant association between functional annotations related to the olfactory system and QTL that were suggestively associated with endo-parasite phenotypes.
Conclusions: A number of novel genomic regions were suggestively associated with endo-parasite phenotypes across the bovine genome and two genomic regions on BTA21 and 25 were associated with antibody response to N. caninum.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

